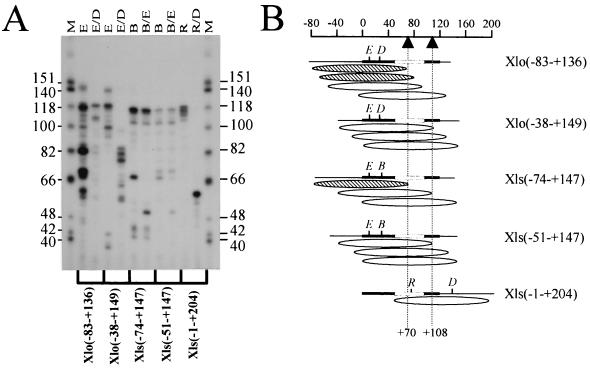
FIG. 2.
Determination of nucleosome translational position on reconstituted X. laevis 5S rRNA gene fragments. (A) The region of DNA in direct association with the histone octamer was determined by digestion of an approximately 146-bp, internally labeled, micrococcal nuclease-resistant fragment with different combinations of restriction enzymes as indicated. The resulting restriction fragments were resolved by denaturing polyacrylamide gel electrophoresis (8% acrylamide and 8.3 M urea in 1× TBE). Lanes M, HinfI-cut φX174 DNA used as a marker (sizes of marker fragments are shown as numbers of nucleotides). The restriction enzymes used are indicated above the gel: E, EaeI; D, DdeI; B, Bsp1286; R, RsaI. (B) Schematic representation of the most predominant nucleosome positions on the five different fragments of the X. laevis oocyte and somatic 5S rRNA genes resulting from the electrophoretic analysis shown in panel A. Due to variations in base composition and thus radioactive labeling, the intensities of the bands in relation to the dATP contents of the fragments were considered in these calculations. The ellipsoids indicate the most abundant positions of the approximately 146-bp micrococcal nuclease-resistant fragments. The thick black lines represent the 5S rRNA coding sequence, and the open boxes indicate the intragenic TFIIIA binding site. Nucleotide positions relative to the transcription start site are indicated on the scale at the top, and the dashed vertical lines indicate positions +70 and +108. The hatched ellipsoids are those nucleosome positions postulated to allow TFIIIA binding, whereas open ellipsoids are those which are thought to be repressive to TFIIIA binding based on the results of You et al. (48).