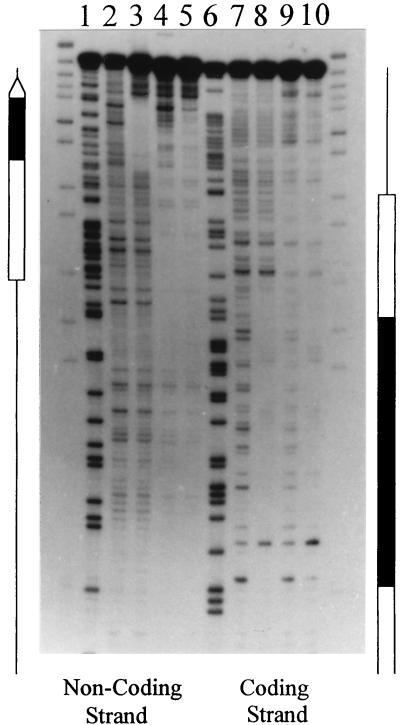
FIG. 5.
DNase I footprint analysis of the complexes formed by nucleosomes and/or TFIIIA on the Xlo(−83→+136) 5S rRNA gene fragment (Fig. 3B). Nucleosomes labeled at the 3′ end of the coding and noncoding strands were incubated in the presence of TFIIIA and subsequently digested with DNase I. The partially digested TFIIIA-nucleosome complexes were purified from free DNA and unbound nucleosomes by native gel electrophoresis (see Materials and Methods). The footprints for the coding and noncoding strands for naked DNA (lanes 2 and 7), TFIIIA-bound DNA (lanes 3 and 8), reconstituted nucleosome (lanes 4 and 9), and TFIIIA-nucleosome complexes (lanes 5 and 10) are shown. Also shown are the Maxam-Gilbert reactions of the labeled DNA (lanes 1 and 6). The 5S rRNA gene is indicated by the open arrow, and the TFIIIA binding site is indicated by a black box.