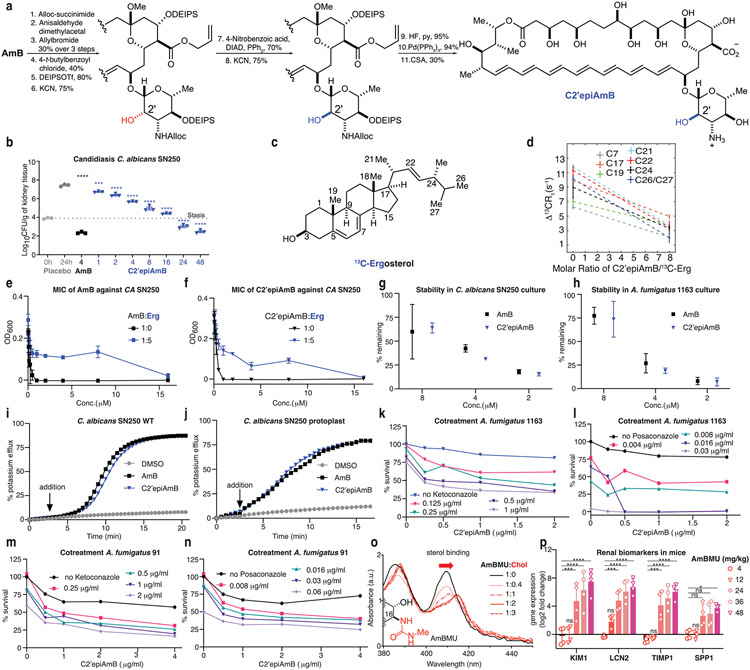
Extended Data Fig. 6 ∣. Mechanistic probing of C2’epiAmB’s decreased potency.
a, Synthesis of C2’epiAmB starting from AmB. b, Evaluations of C2’epiAmB efficacy in disseminated candidiasis mice model infected with C. albicans SN250, 24 h post single IV dose (n = 3 mice/group). Both compounds were administered as 1:2 deoxycholate in D5W to improve aqueous solubility. All dose units are mg/kg. Pairwise comparisons with 24 h placebo group were performed using (one-way ANOVA with Tukey’s multiple comparison test; ***P = 0.0003; ****P < 0.0001. c, Structure and numbering system of skip labeled 13C-erg. Sterol sequestration mechanism of antifungal action is conserved in C2’epiAmB as probed in d, PRE experiments, showing a decrease in the PRE effects of resolved Erg resonances in the presence of C2’epiAmB, indicating a decrease in sterol proximity to doxyl-labeled lipids (samples 40:1 POPC/ 13C-Erg ± 5 mol% 16-DOXYL-PC; Data presented as mean +/− SD of 3 measurements. Error bars indicate uncertainty from the spectrum signal-to-noise ratio.) and decreased efficacy upon pre-complexation with ergosterol for both e, AmB (n = 3 biological replicates/conc.) and f, C2’epiAmB (n = 3 biological replicates/conc.). Both molecules have similar stability profiles in g, C. albicans SN250 (n = 4 biological replicates/conc.) and h, A. fumigatus 1163 cultures (for 8 μM AmB n = 3, for all other n = 4 biological replicates). Both molecules (3 μM) have similar ion channel forming ability and have similar time-delay between compound addition and efflux in i, wild-type C. albicans SN250 and j, instant ion-channel formation in protoplast. Improvement of C2’epiAmB efficacy upon cotreatment with erg biosynthetic inhibitors Ketoconazole and Posaconazole against k, l, A. fumigatus 1163 and m, n, A. fumigatus 91 reveals that the extraction rate-driven kinetic model of efficacy is conserved against moulds. o, Unlike C2’epiAmB, AmBMU, though better tolerated than AmB, retains cholesterol binding and p, causes kidney damage in mice (n = 4 mice/group). Pairwise statistical analyses were performed using two-way ANOVA with Tukey’s multiple comparison test; *P = 0.0422; ***P = 0.0008, ***P = 0.0005, ***P = 0.001, ****P < 0.0001. Results are means ± SD.