Abstract
Root infection in susceptible host species is initiated predominantly in the zone of elongation, whereas the remainder of the root is resistant. Nectria haematococca infection of pea (Pisum sativum) was used as a model to explore possible mechanisms influencing the localization of root infection. The failure to infect the root tip was not due to a failure to induce spore germination at this site, suppression of pathogenicity genes in the fungus, or increased expression of plant defense genes. Instead, exudates from the root tip induce rapid spore germination by a pathway that is independent of nutrient-induced germination. Subsequently, a factor produced during fungal infection and death of border cells at the root apex appears to selectively suppress fungal growth and prevent sporulation. Host-specific mantle formation in response to border cells appears to represent a previously unrecognized form of host-parasite relationship common to diverse species. The dynamics of signal exchange leading to mantle development may play a key role in fostering plant health, by protecting root meristems from pathogenic invasion.
Root-infecting fungal pathogens are a perennial threat to crops worldwide. Crop loss results from direct damage to root systems as well as increased susceptibility to other stresses (Bruehl, 1986). Soilborne fungi that cause disease are dispersed as spores or sclerotia, which remain in a quiescent state until the appearance of a host plant stimulates germination (Katan, 1996). Because the initiation of infection depends on signals that induce spore germination, such signaling comprises a potential target for disease control (Deacon, 1996). Significant progress has been made in defining signal exchange between root exudates and rhizosphere bacteria (VanBrussel et al., 1990; Steele et al., 1999; Endre et al., 2002; Loh et al., 2002a, 2002b; Ferguson and Mathesius, 2003; Hirsch et al., 2003; Limpens and Bisseling, 2003). Rhizobium-legume symbioses appear to share common elements with infection by vesicular arbuscular mycorrhizal fungi, including recognition through flavonoid signals (e.g. Nair et al., 1991; Tsai and Phillips, 1991; Harrison and Dixon, 1993; Hirsch and Kapulnik, 1998; Garcia-Garrido and Ocampo, 2002).
To date, however, our understanding of signal exchange between roots and soilborne pathogenic fungi remains in its infancy (Nelson, 2004). Both seed and root exudates can stimulate spore germination (Curl and Truelove, 1986; Nelson, 1991). Nutrients such as sugars and amino acids leaking from roots have been presumed to serve as nonspecific signals for germination because such compounds are present in root exudates and also can stimulate germination under controlled conditions (Parkinson, 1955; Lockwood, 1988). If it is true that metabolites common to all plants stimulate germination of diverse fungi, then the potential for controlling disease by manipulating the plant's delivery of signals that induce fungal growth may be limited.
Under a variety of conditions, the carbon-rich material collected as exudates is released predominantly at the root apex (e.g. McDougall and Rovira, 1970; Van Egeraat, 1975; Griffin et al., 1976). This material is composed of border cells, mucilage, and soluble compounds released by border cells and/or the root tip (for review, see Hawes et al., 1998, 2000, 2003). The root apex also is the principal source of key signals in root-microbe symbioses (Peters and Long, 1988; Nagahashi and Douds, 2004). The concentration of Bradyrhizobium japonicum nodulation gene inducing flavonoids in the 1-mm root tip of soybean seedlings, in fact, is 10-fold higher than any other part of the plant (Graham, 1991). Yet, paradoxically, the root tip seldom is infected or colonized (Foster et al., 1983; Lagopodi et al., 2002). Instead, infection by microbial pathogens and symbionts generally is initiated just behind the root apex, in the zone of elongation (e.g. Bauer, 1981; Bruehl, 1986; Baluška et al., 1996). This region where newly generated cells from the apical meristem undergo rapid elongation has been defined as the mycorrhizal infection zone to reflect its susceptibility to infection by mycorrhizal fungi (Brunner and Scheidegger, 1992). The region of elongation also is the primary site of infection by fungal pathogens (Fig. 1). The reasons why this site is highly susceptible, while newly generated cells within root tips escape infection, has received little attention.
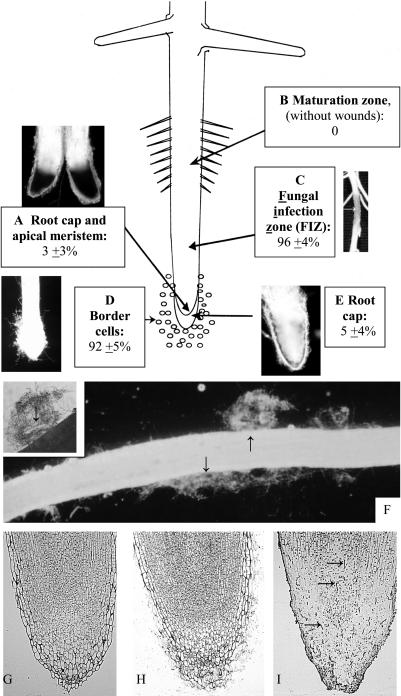
Figure 1.
Localized infection by N. haematococca in the (A) apical and root cap meristem (visualized with trypan blue, which stains dead cells), (B) maturation zone, (C) elongation zone/FIZ, (D) border cells, and (E) root cap. F, Dispersal of border cell mantles along the length of an inoculated root as it grows. Arrows (and inset) indicate clumps of border cells supporting fungal growth. G to I, Cross section of root apex (apical meristem, root cap meristem, root cap body) after removal of border cells from uninoculated control seedlings (G), after removal of border cell mantles from inoculated root with infection into peripheral root cap (H), and after removal of border cell mantles from a root tip infected throughout the root cap and apical meristem (I). Arrows indicate individual fungal hyphae.
Pea (Pisum sativum) has been an important experimental model from the time of Mendel and Darwin, and remains among the world's most important food crops. Nectria haematococca mating population VI (Fusarium solani f. sp. pisi) has served as a long-standing model for aspects of pea root infection (Chi et al., 1964; Cook and Flentje, 1967; deWit-Elshove and Fuchs, 1971; Kraft, 1974; Loria and Lacy, 1979; VanEtten et al., 1980; Stahl et al., 1994). Spores of this fungus, which causes foot and root rot, germinate within 24 h in a 7-mm radius of a germinating seed (Short and Lacy, 1974). Glc and amino acids, which are present in pea root exudate and can induce spore germination, have been proposed to comprise the plant signals triggering germination (Schroth et al., 1963; Ayers and Thornton, 1968; Short and Lacy, 1976). Spore germination also is induced by specific flavonoids and isoflavonoids (Ruan et al., 1995). These metabolites commonly are found in root exudates of legumes (d'Arcy-Lameta, 1986; Maxwell and Phillips, 1990; Cooper and Rao, 1992; Recourt et al., 1992; Wojtaszek et al., 1993; Tsanuo et al., 2003). Flavonoid-induced spore germination is partially blocked by H89, a cAMP-dependent inhibitor of protein kinase A. Root exudate-induced spore germination was found to occur by two independent mechanisms, the flavonoid-responsive pathway that is inhibited by H89 and a nutrient-responsive pathway that is impervious to H89 (Ruan et al., 1995). This discovery provides an experimental framework to compare the role of nonspecific germination stimulators like sugars and amino acids versus specific compounds released by roots of host plants, in the early stages of root-microbe recognition and response.
Previously, we presented evidence consistent with the hypothesis that root tips are protected from fungal infection by the production of root border cells (Gunawardena and Hawes, 2002; Woo et al., 2004). In this study, we used N. haematococca and pea as models to examine the possibility that differential spore germination, pathogenicity gene expression, or defense gene expression is responsible for localized root infection. Instead, we observed that infection by N. haematococca induces border cell death in correlation with development of a factor that selectively inhibits fungal growth.
RESULTS
Despite Formation of a Mantle of Hyphae at the Root Tip, Infection of Host Roots by Pathogenic Fungi Occurs in the Zone of Elongation and Root Tips Remain Uninfected
Radicles (length 2.5 cm) were inoculated uniformly with spores and transferred to cellophane growth pouches. Visible lesions developed 3 d after inoculation with N. haematococca strain 34-18 (Funnell et al., 2001), containing the conditionally dispensable (CD) chromosome with pea pathogenicity genes (PEP; Han et al., 2001; Funnell et al., 2002). Under the conditions used, infection at the cotyledonary attachment site (foot root) occurred in 15% ± 8% of inoculated seedlings (data not shown). On the root, visible lesions that encompassed and killed cells within the root cap and apical meristem were rare (Fig. 1A). Lesions developed in the maturation zone only when it was wounded (Fig. 1B). On most roots, a tan lesion developed within the site where the elongation zone had been, at the time of inoculation (Fig. 1C). This site, which corresponds to the mycorrhizal infection zone (Brunner and Scheidegger, 1992), therefore is referred to as the fungal infection zone (FIZ) to reflect its susceptibility to pathogenic as well as symbiotic fungi. The presence of lesions in the FIZ was not correlated with negative effects on growth and development; 1 week after inoculation, mean root length, shoot height, and number of lateral roots for 225 control and 200 infected plants, respectively, were 174 ± 15 and 180 ± 20 mm, 52 ± 14 and 54 ± 9 mm, and 21 ± 4 and 25 ± 6 per root.
Most plants (>90%) also developed mantles of hyphae supported by growth on border cells (Fig. 1D). Despite being covered in fungal hyphae, visible lesions in the root cap were rare (Fig. 1E). When lesions developed at the cap periphery, cap turnover was induced and mantles detached and were dispersed along the length of the root as it grew (Fig. 1F, arrows); at higher magnification, a nucleus of border cells could be seen within each mantle (Fig. 1F, inset, arrow). In roots with lesions confined to the root cap periphery (as in Fig. 1E), the cellular structure of the root cap (Fig. 1H) was comparable to that of control root caps (Fig. 1G). By contrast, visible lesions throughout the apex (as in Fig. 1A) were associated with disintegration of cellular organization, and fungal hyphae could be seen throughout the tissue (Fig. 1I, arrows).
The same results were obtained with four pea cultivars (Early Frosty, Lincoln, Progress 9, and Green Arrow) as well as alfalfa (Medicago sativa) inoculated with N. haematococca 34-18, and on cv Little Marvel in response to other pea pathogens, including Phoma pinodella (L.K. Jones) T409, Mycosphaerella pinoides (Berk. & Bloxam) T417, Thielaviopsis basicola, and nine different isolates of N. haematococca (data not shown). In each case, nearly all roots developed lesions in the FIZ but not within the root tip, despite the presence of mantles on root caps (as in Fig. 1D). In response to inoculation of Little Marvel with N. haematococca strains 94-2-4 and 44-100, which lack the CD chromosome, mantles that formed were smaller than those produced by other strains, and 50% ± 7% of inoculated seedlings failed to develop a visible lesion in the FIZ. The non-pea pathogenic strain of T. basicola did not induce any lesions or mantles on roots of pea seedlings, and no mantles or lesions developed in response to N. haematococca 34-18 on roots of nonhost species, including wheat, cotton, soybean, cucumber, pumpkin, or corn (data not shown).
Root Tip Escape from Infection: Spore Germination at the Root Tip
Exudates Stimulate Rather Than Inhibit Spore Germination
One hypothesis to explain how fungus-covered root tips avoid infection is that border cells and associated mucilages form a physical barrier to prevent contact between infective propagules and the root cap periphery. No evidence for physical exclusion was found (Fig. 2). A second possibility, that spores fail to germinate at the root tip region, was explored. Spores (105/mL) added to pea roots began to germinate within 1 h, and >90% had germinated within 2 h. Washed border cells alone were equally effective (Table I). The presence of the CD chromosome did not influence the rate of germination. No significant germination occurred in water over a 6-h period.
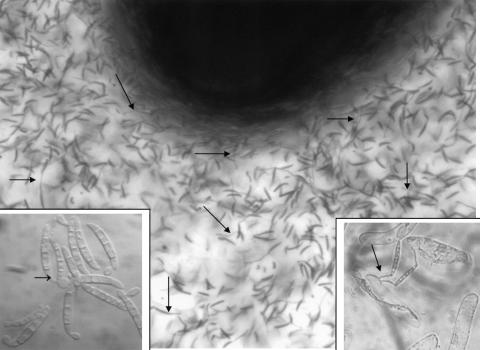
Figure 2.
Germination of N. haematococca spores in situ, in response to root cap and border cell exudates. After immersion of root tips into a suspension of spores at 105/mL, spores start to germinate within 1 h (arrows). Inset, Left, At 106 spores/mL, chlamydospore formation occurs. Inset, Right, Fungal hyphae grow around border cells without apparent penetration.
Table I.
Stimulation of spore germination by pea root cap and border cell exudatesa
| N. haematococca Strainb | Root Exudate | Border Cells | Waterc |
|---|---|---|---|
| 34-18 (+CD chromosome) | 89% ± 5% | 87% ± 5% | <1% |
| 77-13-4 (+CD chromosome) | 94% ± 4% | 90 ± 4 | <1% |
| 94-2-4 (−CD chromosome) | 91% ± 4% | 89% ± 9% | <1% |
| 44-100 (−CD chromosome) | 100% | 78% ± 6% | <1% |
For strain 34-18, the data represent mean values from five different trials, each of which contained two replicates. For other experiments, the values represent mean values from two different trials, each of which contained two replicates. Samples were processed as described in “Materials and Methods.”
Isolates with and without the CD chromosome with PEP genes were added at a final concentration of 105/mL and incubated for 2 h.
No significant germination occurred over a 6-h period of observation.
Bulk exudate or border cells alone, when separated from the root, induced maximum spore germination (Table II). At 104 spores/mL, 100% germination occurred within 1 h. Germination was reduced at higher spore concentrations, presumably due to endogenous germination inhibitors (e.g. Bruehl, 1986). At 106 spores/mL, germination decreased to 72% ± 8%, and chlamydospore formation was induced (Fig. 2, inset left); adding Glc (2.5 mg/mL) to the medium did not increase germination or suppress chlamydospore formation (data not shown). At 107 spores/mL, no germination occurred (Table II).
Table II.
Dosage response for root cap and border cell exudate stimulated germination of N. haematococca 34-18 spores
The data represent mean values from three different trials each of which contained two replicates.
| Treatment
|
Percent Spore Germination after 2.5 h
|
|||
|---|---|---|---|---|
| 105 | 2 × 105 | 106 | 107 Spores/mL | |
| Water | 4 ± 2 | 1 ± 2 | 0 | 0 |
| Exudate (2.5 roots/mL) | 98 ± 2 | 99 ± 1 | 72 ± 4 | 0 |
| Exudate (5.0 roots/mL) | 99 ± 1 | 99 ± 1 | 95 ± 4 | 0 |
| Exudate (10 roots/mL) | 98 ± 2 | 100 | 98 ± 2 | 0 |
The Nature of the Spore Germination Stimulant
The germination stimulation activity was stable to freezing and boiling for 30 min and, in preliminary tests, was retained in the water phase when exudates were extracted with organic solvents (data not shown). One candidate for the activity therefore is amino acids present within root exudates (Bruehl, 1986; Phillips et al., 2004). Total dry weight of root cap and border cell exudates, after separation from the root and border cells, was 17.3 μg per root. Sixteen of 20 protein amino acids plus the non-protein amino acid γ-aminobutyric acid were present, at a total amino acid concentration of >1.2 μg per root. Levels of individual amino acids, in picograms per root, were as follows: Asp, 360; Thr, 101; Ala, 118; Val, 80; Met, 10; Ile, 45; Leu, 60; Tyr, 62; His, 190; Arg, 85; and γ-aminobutyric acid, 140. Ser, Glu, Gly, Cys, Phe, and Trp were present in trace amounts. When protein amino acid ratios were reconstituted in water and mixed with N. haematococca spores, no germination occurred over a 6-h period (Table III). Therefore, amino acids do not appear to account for the rapid germination stimulation by root cap and border cell exudates.
Table III.
Inhibition of root cap and border cell exudate-stimulated spore germination by the protein kinase A inhibitor H89a
| Treatment
|
Percent Germination
|
|
|---|---|---|
| Water | Water + H89b | |
| Amino acid mixturec | 0 | 0 |
| Root cap and border cell exudate | 90% ± 5% | 0 |
| Border cell exudate | 89% ± 6% | 0 |
| Glc-Asn medium | 80% ± 8% | 78% ± 9% |
| M100 minimal medium | 84% ± 5% | 87% ± 4% |
Spores (105/mL) from N. haematococca strain 34-18 were incubated for 2 h in exudate (2.5 roots/mL equivalent), and germination was measured by direct counts. Border cell exudate reflects samples of spores incubated with washed border cells in which the only exudate present is that released from the border cells during the test period.
H89 was added to a final concentration of 100 μm. No germination occurred over a 24 h period of observation.
The amino acid mixture consisted of commercial samples of primary amino acids, each reconstituted to the same concentration found to be present in pea root cap and border cell exudates. The data represent mean values from five different trials, each of which contained two replicates. No germination occurred in the amino acid samples after a 6-h period of observation.
To determine whether exudate-induced germination occurs via the nutrient- or the flavonoid-responsive pathway (Ruan et al., 1995), the protein kinase A inhibitor H89 was used. When assayed in growth media, Glc-Asn, or minimal medium with Glc as the only carbon source (M100 minimal medium), germination was unaffected by H89 (Table III). By contrast, H89 completely inhibited exudate-induced spore germination for at least 24 h, the longest time period evaluated (Table III). Staining with the vital stain fluorescein diacetate after 24 h exposure to H89 was indistinguishable from that of untreated spores, and when removed from H89 and plated onto growth medium, the conidia germinated and grew normally; this indicated that the inhibition of germination was not due to loss of spore viability (data not shown).
Root Tip Escape from Infection: Expression of N. haematococca PEP Genes and Plant Defense Genes during Mantle Formation at the Root Tip
Expression of Fungal Pathogenicity Genes
One hypothesis to explain how root tips avoid infection is that hyphae fail to express PEP genes when growing on border cells. A strain expressing uidA encoding β-glucuronidase (GUS) under the control of the pisatin demethylase (PDA) promoter was used as a reporter system for PEP gene expression (Straney and VanEtten, 1994). This promoter is induced during infection of pea and in response to pisatin (Fig. 3A). Incipient mantles were visible on root tips within 24 h (Fig. 3B). At the same time, pisatin (15 μg/g tissue dry weight) was produced in the FIZ, but none was detectable in the root tip (data not shown). Despite the absence of detectable pisatin production, expression of PDA-GUS was visible as blue hyphae within the mantles within 24 h (Fig. 3B, arrow). In some cases, mantles failed to detach and PDA-GUS expression was maintained over several days (Fig. 3C). Expression of other PEP genes also was induced within 24 h after inoculation of the root tip (Fig. 3D) and remained elevated for at least 60 h (data not shown).
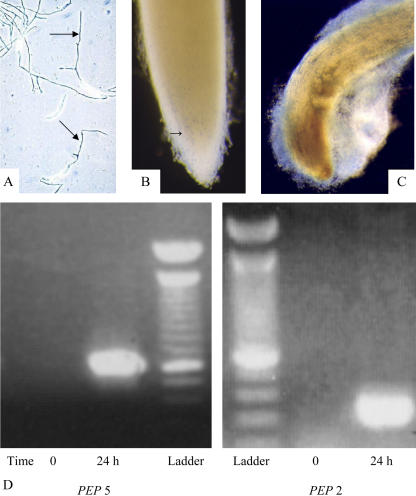
Figure 3.
Expression of PEP genes during early stages of mantle development. A, Expression of PDA-GUS in germinating spores exposed to pisatin; B, expression of PDA-GUS in border cell mantles 24 h post inoculation; C, expression of PDA-GUS in older mantle surrounding infected root tip; and D, expression of PEP5 (left) and PEP2 (right) genes of N. haematococca at time 0 and at 24 h after inoculation. Two-day-old seedlings containing a full set of border cells were inoculated with 106 spores of N. haematococca, and RNA from root tips was recovered at specific time points after inoculation, as described in “Materials and Methods.” A 100-bp ladder was used to establish agreement between expected size and actual size of fragments obtained by amplification of each gene.
Expression of Plant Defense Genes
An alternative mechanism for protecting the root tip is the induction of defense pathways. In a previous study, when seedlings were inoculated with spores from N. haematococca 34-18, chitinase expression in the FIZ increased within 24 h and remained elevated through 72 h, but no such increase in expression occurred in the root tip (Gunawardena and Hawes, 2002). Because root tips rarely develop lesions, the defense gene elicitor salicylic acid (SA) was used in this study to examine defense responses in the root tip. At a concentration of 5 or 10 mm SA, chitinase expression in the FIZ was found to be comparable to that which was previously shown to occur in response to infection; thus, expression was induced by >3-fold in the FIZ after 24 h and remained elevated through 72 h (data not shown). Root growth in response to 5 or 10 mm SA, under the same conditions was inhibited by 46% ± 6% (5 mm) and 64% ± 5% (10 mm). As in roots inoculated with fungal spores, however, no increased chitinase expression was observed in the root tip after 24 h exposure to 5 or 10 mm SA. Only when earlier stages were examined was increased expression found to occur in the root tip, with a transient increase at 3 to 6 h (Fig. 4A). Despite continuous exposure to levels of SA that induced continuous defense expression in the FIZ, expression in the root tip returned to control levels within 8 h and remained low throughout a 72-h period of observation. These data suggest that molecular responses to stress in the root tip are markedly distinct from those in the FIZ.
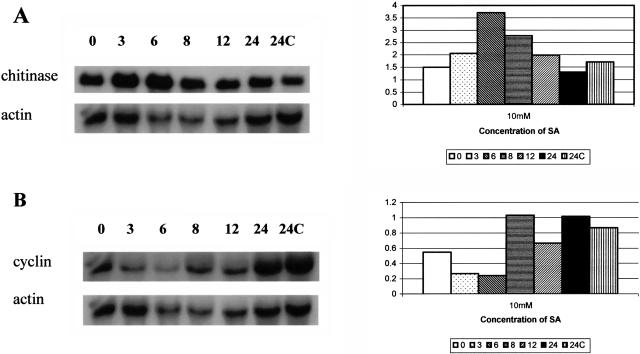
Figure 4.
Inverse relationship between expression of defense genes and cell cycle genes in root tips treated with SA at a level that inhibits root growth. Relative levels of (A) chitinase mRNA and (B) cyclin mRNA were normalized to actin as a control. Seedlings were grown in cellophane pouches in which SA was added at a concentration that resulted in approximately 50% inhibition of root growth; root lengths were measured nondestructively as by Bauer (1981) before harvesting root tips for RNA-blot analysis, as by Gunawardena and Hawes (2002).
Expression of Cell Cycle Genes
The induction of defense pathways in some tissues is inversely correlated with expression of cell cycle-associated genes (Logemann et al., 1995; Sommssich and Hahlbrock, 1998). To examine the potential impact of defense gene expression on cell cycle in root meristems, total RNA isolated from root tips was probed with four cell cycle-related genes during the early stages of SA treatment when chitinase expression is induced. Histone 3, histone 4, and cdk expression was unaffected by SA (data not shown), but cyclin expression was reduced by >2-fold after 3 h exposure to 10 mm SA (Fig. 4B). The reduced expression of cyclin was transient and inversely correlated with chitinase. Thus, cyclin expression decreased as chitinase increased, and both returned to normal at 8 h (Fig. 4, A and B). The same relationship occurred at 5 mm SA (data not shown). The failure to maintain defense pathways during mantle formation, even under stress sufficiently severe to inhibit growth, would be predicted to leave the root tip especially vulnerable to infection.
Root Tip Escape from Infection: Development of a Fungal Growth Inhibitor
Border Cell Death during Mantle Formation
Unlike in root lesions or culture medium where sporulation begins within 24 h, fungus in mantles grew solely as hyphae and failed to sporulate even after 10 to 14 d (Fig. 1, D and F). Mantle development was examined in detail to explore cell-cell interactions between plant and pathogen. Within 18 h after inoculation, border cell death occurred in a dosage-dependent manner in response to pea pathogens, including N. haematococca, P. pinodella, and T. basicola (Table IV). In N. haematococca isolates 94-2-4 and 44-100 lacking the CD chromosome, a 10-fold increase in inoculum was required for border cell death to occur, and no significant cell death occurred in response to an isolate of T. basicola that is not pathogenic on pea (Table IV). The mechanism of border cell death was explored because direct penetration of border cells by N. haematococca was not observed. Most appeared to grow around the cells and then spread to the other areas (Fig. 2, inset right). The possibility that a toxic factor is secreted by the fungus was assessed by challenging populations of border cells with exudate collected from roots after inoculation with N. haematococca or from culture filtrates of N. haematococca grown in M100 medium. No evidence was found for a toxin that by itself, in the absence of the fungus, killed border cells. Contact between border cells and fungal cells appears to be required for border cell death to proceed.
Table IV.
Loss of viability in border cells from pea seedlings inoculated with spores of pathogenic fungi
For each concentration of spores, border cells from three seedlings were collected and viability was measured by staining with the vital stain fluorescein diacetate, as described in “Materials and Methods.” Data represent mean values obtained from at least three independent trials.
| Treatment
|
Percentage of Control Border Cell Viability
|
||
|---|---|---|---|
| 105 | 106 | 107 Spores/mL | |
| N. haematococca 34-18 | |||
| 4 h (agar) | 100% | 100% | 100% |
| 12 h (agar) | 100% | 100% | 100% |
| 18 h (agar) | 100% | 50% | 16% |
| 18 h (pouches) | 90% | 62% | 31% |
| N. haematococca 44-100 | |||
| 18 h | 100% | 100% | 40% |
| N. haematococca 94-2-4 | |||
| 18 h | 100% | 100% | 44% |
| P. pinodella | |||
| 18 h | 88% | 58% | 28% |
| T. basicola (pea pathogenic) | |||
| 18 h | 89% | 64% | 37% |
| T. basicola (non-pea pathogenic) | |||
| 18 h | 100% | 100% | 100% |
Production of a Fungal Growth Inhibitor
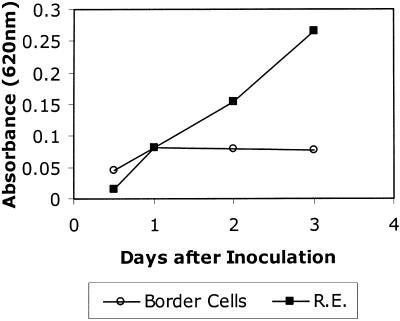
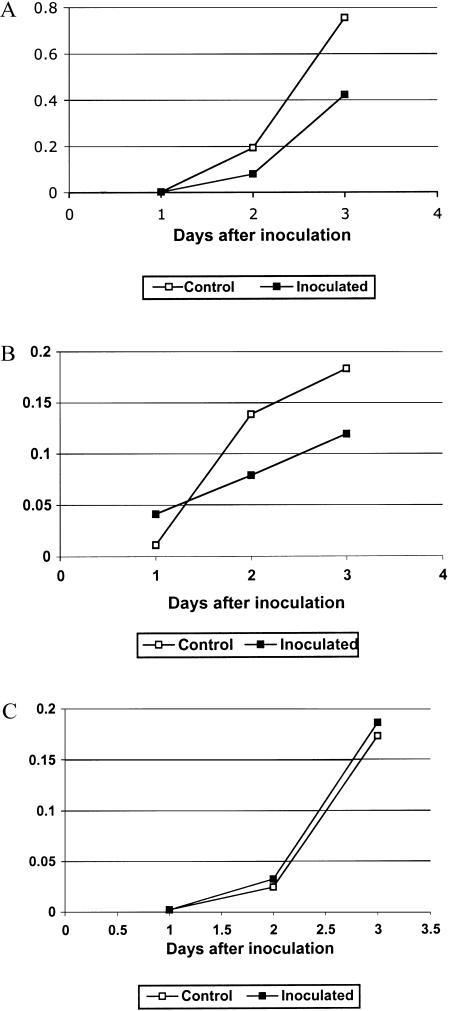
Inhibition of fungal growth occurred in response to cocultivation with border cells (Fig. 5). Whereas growth of fungus on bulk root cap exudates was linear over the course of several days, growth on border cells ceased after 1 d. To determine whether nutrients were limiting or an inhibitor was produced, exudate was collected from control seedlings or from seedlings inoculated with N. haematococca and differences in available sugar were corrected by adding Glc. At 2 to 3 d after inoculation, a significantly higher rate of fungal growth occurred in exudate from control versus inoculated roots (Fig. 6A). When the concentration of exudate from inoculated seedlings was reduced by one-half, twice as much growth occurred (P < 0.015), but no such increase occurred when exudate from control roots was diluted (P value 0.237; Table V). The fungal growth inhibitor was effective against other N. haematococca isolates, with or without the CD chromosome (Fig. 6B). By contrast, M. pinoides grew equally well in exudates of control seedlings and those inoculated with N. haematococca, suggesting that the inhibitor is selective in its effects on fungal growth (Fig. 6C).
Figure 5.
Differential growth of N. haematococca in root exudates (R.E.) compared with border cells. Spores were added to samples of border cells or bulk root cap exudate, and growth was monitored over the course of 3 d, as described in “Materials and Methods.” Values represent means from four independent experiments, and ses were less than 10% of the mean.
Figure 6.
Inhibition of growth of N. haematococca in root exudates of inoculated seedlings. A, Spores (105/mL) of N. haematococca 34-18 were added to samples of root exudate collected from control or inoculated seedlings, and growth was monitored over the course of 3 d, as described in “Materials and Methods.” The difference in the growth was statistically significant for the growth in the second (P value 0.009) and the third day (P value 0.006) but not for the first day (P value 0.745). B, Growth of N. haematococca 94-2-4 in exudate from seedlings inoculated with strain 34-18 and collected as described in “Materials and Methods.” Values for exudate from control or inoculated seedlings at day 2 and day 3 were statistically distinct at P < 0.05. C, Growth of M. pinoides in exudate from seedlings inoculated with N. haematococca strain 34-18 and collected as described in “Materials and Methods.” Values were not statistically distinct at P < 0.05.
Table V.
Presence of a fungal growth inhibitor in root cap and border cell exudate from inoculated roots: increased growth in response to dilution of exudatea
| Exudate from
|
Absorbance (620 nm)
|
||||
|---|---|---|---|---|---|
| Trial
|
Mean
|
P Value
|
|||
| 1 | 2 | 3 | |||
| Inoculated seedlings, undiluted | 0.09 | 0.07 | 0.075 | 0.078 | 0.015 |
| Inoculated seedlings, 50% | 0.17 | 0.13 | 0.14 | 0.146 | |
| Control seedlings, undiluted | 0.18 | 0.175 | 0.26 | 0.205 | 0.237 |
| Control seedlings, 50% | 0.25 | 0.23 | 0.255 | 0.245 | |
Root cap/border cell exudate from inoculated seedlings and control seedlings obtained, as described in “Materials and Methods,” was used at full strength or diluted by one-half by addition of water.
DISCUSSION
The term rhizosphere was coined by Hiltner (1904) based on his recognition that microorganisms grow in the immediate vicinity of roots and that these microbial communities impact plant health. Localized microbial growth in the rhizosphere occurs because nutrient-rich materials are released from roots into the extracellular environment (Rovira, 1956, 1959; Harmsen and Jager, 1962). Intensive research therefore has been dedicated to defining microbial traits that foster their ability to colonize this habitat (Rovira, 1991; Gilbert et al., 1996; Dunn et al., 2003). Surprisingly, however, despite a long-standing recognition that the plant genotype is at least as important as microbial genotype in controlling rhizosphere community structure (e.g. Atkinson et al., 1975; Curl and Truelove, 1986), there never has been a corresponding emphasis on defining plant genes that control the composition and release of root exudates (O'Connell et al., 1996; Smith et al., 1999).
In part, this omission has been based on a presumption that most exudation involves a continuous global release of sugars, amino acids, and other nutrients along the surface of the root, and that this mixture would be predicted to support growth of microbial populations in a nonspecific manner (e.g. Bar-Yosef, 1996). Available evidence from root systems grown in systems ranging from culture plates to hydroponics to native soil has never been in support of such a model (for review, see Hawes et al., 2003). In most plant species, the bulk of carbon-rich material released from roots is programmed to separate at the apex of the root (e.g. McDougall and Rovira, 1970; Van Egeraat, 1975). Most of the material, moreover, is packaged in living cells (Knudson, 1919; Griffin et al., 1976; Vermeer and McCully, 1982). As a root moves into regions where specific microorganisms are present, this mixture is the first source of signals to contact dormant propagules in the soil. As with any living plant cell, however, the material contained within border cells is only accessible to microorganisms that carry the genes needed to recognize and respond to those signals (Hawes et al., 1998, 2000, 2003). The capacity of fungi and bacteria to recognize and respond to other substrates in the form of mucilage, cell wall breakdown products, and other materials that detach along with border cells also appears to be species and genotype specific (Goldberg et al., 1989; Teplitski et al., 2000; Knee et al., 2001).
In this study, we observed strong host selectivity in germination and mycelial growth in response to exudates from the root cap and border cells. There was no mantle formation without a pathogenic fungus and susceptible host. Most important, spore germination occurred very rapidly by the same pathway as flavonoid-induced germination, and when this pathway was blocked, spores remained in a quiescent state. Thus, availability of nonspecific metabolites like amino acids and sugars, shown previously to induce germination of fungal spores, is not sufficient to induce germination and growth when the flavonoid-responsive pathway is blocked. Manipulation of specific genes in plant roots to control the release of factors controlling spore germination therefore might be predicted to protect roots from infection by soilborne pathogens without necessarily compromising symbiotic infection by beneficial bacteria and fungi. If the factor is a flavonoid, for example, then localized expression of enzymes that modify their activity by glycosylation could be used to test their role in germination (Hsieh and Graham, 2001). Known flavonoids and isoflavonoids in the aglyone form are soluble in organic solvents, but the germination stimulant from pea did not exhibit this property. However, such products frequently occur in root exudates as water-soluble conjugates (e.g. Graham, 1991; Recourt et al., 1992). Moreover, H89 can influence activity of multiple targets, and it is possible that more than one pathway may be involved in synthesis of specific signals that induce spore germination (Syin et al., 2001; Leemhuis et al., 2002).
Border cells appear to be a primary, if not the only, source of the germination stimulant. Thus, exudates released from border cells alone induced germination as effectively as the mixture of bulk root cap and border cell exudates. Soilborne pathogenic fungi can interact with border cells by several mechanisms (for review, see Hawes et al., 1998, 2000). Zoospores of pathogenic Pythium spp. exhibit genotype-specific chemotaxis to the border cells of host species, then penetrate and cause cell death within minutes (Goldberg et al., 1989). Host-specific toxins from pathogenic Cochliobolus spp. kill border cells only in genotypes of oats and corn carrying specific genes for susceptibility to the fungi (Hawes and Wheeler, 1982; Hawes, 1983). Border cells of corn develop papillae, in a genotype-specific manner, in response to infection by Colletotrichum graminicola (Sherwood, 1987). In this study, an additional mechanism appears to operate, in which cell-cell contact is needed for border cell death and also for production of a substance that modulates fungal growth. Border cells of bean cultivars undergo hypersensitive cell death in response to Pseudomonas phaseolicola strains carrying appropriate avirulence genes (M.C. Hawes, unpublished data), and a similar mechanism involving elicitation of programmed cell death might operate in response to fungal pathogenesis.
Ectomycorrhizal fungi and Fusarium oxysporum previously have been reported to associate and grow among border cells at the root tip of host species during initial interaction with host roots (Fusconi, 1983; Horan et al., 1988; Brunner and Scheidegger, 1992; Turlier et al., 1994; Rodriguez-Galvez and Mendgen, 1995; Kroes et al., 1998). This was presumed to involve a saprophytic interaction based on the incorrect notion that border cells die before detachment from the root, so no information is available about the mechanism of cell death. Nevertheless, the data suggest that growth on border cells in early stages of contact is a phenomenon common to diverse fungi. Eighty percent of land plants develop symbiotic associations with arbuscular mycorrhizal fungi, and this capacity is tightly correlated with production of border cells; the small number of species that fail to develop such associations, including Arabidopsis (Arabidopsis thaliana), are the same species that fail to produce populations of living border cells (Niemira et al., 1996; Arriola et al., 1997). A recent study has documented that a product from border cells stimulates branching in the arbuscular mycorrhizal fungus Gigaspora gigantea, which might in part explain this correlation between production of border cells and susceptibility to mycorrhizal infection (Nagahashi and Douds, 2004).
CONCLUSION
In summary, even under conditions highly conducive to disease (i.e. succulent young roots inoculated directly with thousands of spores and maintained at warm temperatures in cellophane pouches), lesion development on unwounded roots is tightly localized within the region of elongation. Our results are consistent with the hypothesis that the root tip escapes infection by a multiphasic guarding effect of border cells that release signals to stimulate spore germination then modulate hyphal growth. The result is to allow the establishment of a stable relationship in which the fungus survives without damaging root function. Further research is needed to examine the possibility that having a pathogen like N. haematococca established in the root-rhizosphere environment of its host, without causing disease, may actually provide ecological benefits to the plant. As with symbiotic bacteria and fungi, the capacity to live in close proximity and utilize resources of the plant without causing lesions that inhibit growth and development could be advantageous to soilborne fungal pathogens.
MATERIALS AND METHODS
Seedling Germination and Maintenance of Nectria haematococca Cultures
Pea (Pisum sativum) seeds were surface sterilized by immersion in 95% ethanol for 10 min followed by 30 min in 5% sodium hypochlorite (Brigham et al., 1998). Throughout a 6-h period of imbibition in sterile water, seeds that floated were discarded. Remaining seeds were spread on germination paper overlaid on 1% water agar and incubated at 24°C for 2 d in the dark.
Nectria haematococca (Berk. & Br) mating population VI, isolate 34-18, containing the CD chromosome with PEP genes was used in most experiments (Funnell et al., 2001, 2002; Han et al., 2001). Other isolates were N. haematococca T2, T8, T23, T406, T547, SUF1111, and 77-13-4 that cause foot and root rot in pea (Delserone et al., 1999; Funnell et al., 2001); strains 94-2-4 and 44-100, without the CD chromosome, which do not cause foot and root rot (Wassman and Van Etten, 1996; H.D. VanEtten, unpublished data); and pea pathogens Phoma pinodella (L.K. Jones) T409, Mycosphaerella pinoides (Berk. & Bloxam) T417, and Thielaviopsis basicola D127 (Delserone et al., 1999; H.D. VanEtten, unpublished data). A non-pea pathogenic isolate of T. basicola, Smith Henson 2, a pathogen of tobacco, was used as a control. Isolates were maintained on V8 juice agar plates (medium 29; Stevens, 1974). For T. basicola cultures, 0.1% Glc was added to the medium. The cultures were at least a week old when spores were harvested. The conidial suspension was centrifuged at 3,500g for 1 min, washed once with sterilized water, and resuspended in water. Spores were enumerated using a hemocytometer, and the concentrations were adjusted to the desired values by dilution with water.
Root Infection Assays
Four seedlings with a radicle length of 2.5 cm were inoculated with spores, then inserted into a cellophane growth pouch (Mega International, Minneapolis) containing 15 mL of water, covered to prevent drying, and incubated at 24°C in cellophane growth pouches for up to 10 d. Infection was evaluated by direct observation and/or plating onto medium to assess fungal growth, as described (Gunawardena and Hawes, 2002). For mantle formation and infection by different fungal isolates, at least 15 seedlings for each treatment in at least three trials were included for each combination.
Collection of Root Cap and Border Cell Exudates
Root tips (approximately 1–2 mm of the root apex, including the root cap and apical meristem) of intact 2-d-old seedlings with a radicle length of 2.5 cm were immersed in 1 mL of water and washed by gently pipetting with water, as described (Zhu et al., 1997; Brigham et al., 1998). Border cells (approximately 4,000 cells per root tip) were removed from the collected exudates by centrifugation at 3,500g for 1 min and washed twice to remove all extracellular material. Root cap and border cell exudate refers to the supernatant component obtained after this centrifugation step.
To collect exudates and culture filtrates for border cell toxin assays, two sets of 50 seedlings (radicle length 2.5 cm) were placed on 1% water agar overlaid with germination paper. Each seedling was inoculated with 50 μL of water containing 0 or 106 spores of N. haematococca applied to the radicle. The trays were covered and incubated at 24°C in the dark for 24 h. Root tips of the inoculated versus control seedlings were immersed in 1 mL of water, and the tips were gently washed by pipetting water onto the tip. The resulting suspensions were passed through a 0.2-μm filter, and a 500-μL sample was distributed into each of six wells of a 24-well plate. These were inoculated with 106 spores of N. haematococca or water per well. The plate was sealed with parafilm and shaken at room temperature for 24 h. The contents of the wells were pooled, centrifuged, and the supernatant filtered (0.2 μm) before assaying for toxicity. Culture filtrates were harvested from fungal cultures grown 7 d in M100 minimal medium inoculated with 106 spores/mL.
To collect exudates for fungal growth assays, control or inoculated seedlings were processed as described above. After 24 h at 24°C, each seedling was inserted into a 5-mL glass test tube with the root tip submerged in water, and incubated in the dark at 24°C. After 24 h, the liquid was recovered, centrifuged at 3,900g for 5 min, and the supernatant frozen in 100- to 150-mL aliquots, lyophilized, and dissolved in 500 μL of water. Carbohydrate concentration in exudates was measured (Dische, 1962), based on mean values of five independent batches of collected root exudates. Protein concentration was measured (catalog no. 500-0006; Bio-Rad, Hercules, CA). The carbohydrate concentration in root tip exudates of inoculated seedlings contained less carbohydrate (180 ± 15 μg/100 μL) than that of the noninoculated seedlings (325 ± 25 μg/100 μL), and Glc was added to equalize available sugar in the two samples. Protein in exudates from inoculated seedlings was increased to 145 ± 10 from 95 ± 10 in exudates from uninoculated controls.
Spore Germination Assays
To evaluate germination, spores were added to exudate, border cells, or culture medium in a 24-well microtiter plate in a final volume of 400 μL. Replicate 10-μL samples were spotted onto a glass slide, and percent germination of 200 to 300 spores was recorded. For all experiments, water was used as a control. To evaluate dosage response, exudates from 0, 2.5, 5, and 10 tips were contained in 400 μL of water and mixed with varying concentrations of spores. H89 (catalog no. 371963; Calbiochem, San Diego) was dissolved in water and added at a final concentration of 100 μm (Ruan et al., 1995).
Amino Acid Analysis and Assay
Free amino acid analysis was conducted with a Dionex model MBF/SS amino acid/peptide analyzer (Sunnyvale, CA) equipped with a Gilson Spectra/GLo fluorometer (Middleton, WI) with wavelength of excitation at 360 nm and emission at 455 nm. Exudate from Little Marvel pea (10.9 mg dry weight, collected from 650 root tips and separated from border cells as described above) was loaded onto a 0.4- × 12.0-cm column packed with DC-5A cation exchange resin (Dionex) by a 10-μL calibrated injection loop. The separated amino acids were subject to detection following post-column derivatization with o-phthaldialdehyde (OPA; Roth, 1971). Peak identification and integration of peak areas was performed with Shimadzu C-R3A Chromatopac data processor/recorder (Columbia, MD). The peaks were identified by absolute retention time and quantified by peak area using a standard curve generated from standards of known concentration by the least squares method of linear regression. Amino acids were sequentially eluted from the column using six buffers of increasing pH in a constant 0.2 m solution of Na+ cations. These buffers contained 2.0 g of NaOH (Gallard-Schlesinger AnalaR grade; Plainview, NY), 8.76 g of NaCl (Gallard-Schlesinger AnalaR grade), 0.25 g of NaEDTA, and 1.0 mL of phenol (Mallinckrodt, Hazelwood, MO) per liter. The sample buffer was titrated to pH 1.0, eluent A to pH 3.1, eluent B to pH 3.5, and eluent C to pH 4.2 using formic acid. Eluent D was titrated to pH 7.1 with H3PO4 (Fisher Scientific, Loughborough, UK), and eluent E was brought to pH 10.0 with 3.5 g of H3BO4 (Gallard-Schlesinger AnalaR grade) and two pellets of NaOH. An additional regenerant buffer, eluent F, contained 4.0 g of NaOH, 5.84 g of NaCl, 0.25 g of Na2EDTA, and 1.0 mL of phenol per liter.
For primary amino acids, eluents were pumped through the column at a flow rate of 15.0 mL/h (column temperature 43°C), and 20-μL aliquots of exudate were injected into the eluent flow. After leaving the column, the amino acid containing eluent was mixed with OPA/2-mercaptoethanol complex, 9.0 g of OPA dissolved completely in 2 to 3 mL of methanol, 12.0 mL of 2-mercaptoethanol, 61.8 g of H3BO3, 48.0 g of KOH, and 3.0 mL of Brij-35 per liter solution, at a flow rate of 5 mL/h. The eluent-OPA/2-mercaptoethanol mixture entered delay coil B, where fluorophor formation occurred (1 min, room temperature) prior to measurement in the fluorometer.
Border Cell Viability during Mantle Formation
Border cell toxin bioassays were carried out as described (Hawes and Wheeler, 1982; Hawes, 1983). At indicated intervals after inoculation of roots, border cells were washed into 1 mL of water. Viability was evaluated using the vital stain fluorescein diacetate. Each trial consisted of at least two independent experiments, each of which included at least two replicates containing three individual samples. To assess for the presence of an extracellular toxin developing during infection, a 500-μL sample of the filtered suspensions from inoculated or uninoculated seedlings was applied to two wells of a 24-well plate. Washed border cells from five roots (approximately 20,000 total cells) were introduced into each well. The plate was covered in aluminum foil and gently shaken for 6 h at room temperature. A sample of the suspension was diluted to 100 μL, and 10 μL of this suspension was used to assess the viability; values are expressed as percentage of control viability of border cells maintained in water. Viability of control border cells was 89% ± 8% at the start and was reduced to 70% ± 15% after 24 h. Three independent trials, each of which contained two replicates, were carried out.
To evaluate toxin production in fungal culture, four seedlings were inserted into sterile growth pouches containing 15 mL of the fungal culture filtrate (M100+), M100 medium (M100), or water. Two replicates for each treatment were set up for each trial. The bags were placed in a plastic container and were loosely wrapped with Saran Wrap (Dow Chemical, Midland, MI) to prevent drying. The containers were placed in a 24°C incubator in the dark. At 24 and 48 h after initiation of the experiment, the four seedlings in each treatment were pooled, washed, and the viability of the border cells evaluated by microscopic observation using fluorescein diacetate as the vital stain.
Fungal Growth in Root Cap and Border Cell Exudates
Exudates (100 μL at 0.2 mg) from control or inoculated seedlings were mixed with 500 spores in wells of a 96-well microtiter plate. Blank wells with exudate or border cells without fungal inoculum served as controls. Fungal growth was assessed by measuring A620 using a microwell plate reader (Titertek Multiscan MCC/340; Huntsville, AL) and subtracting the blank well values. For each value, the spectrophotometric data were confirmed by visual inspection using a dissecting microscope (SV8; Zeiss, Jena, Germany).
Defense- and Cell Cycle-Related Gene Induction
Root tips and FIZ were excised at intervals after treatment with SA (sodium salt; Sigma Chemical, St. Louis), then frozen and stored at −80°C. Northern analysis was as described (Gunawardena and Hawes, 2002). Probes (below) were labeled with 32P by the random primer method using the Prime It II kit from Stratagene (catalog no. 300385; La Jolla, CA). Hybridization was carried out at 42°C for heterologous probes and at 50°C for homologous probes using the Ultra Hyb hybridization buffer obtained from Ambion (catalog no. 8670; Austin, TX). Density quantification of the signals obtained from the northern analysis was done using the NIH Image software program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). Relative level means the ratio of the density values obtained for the signal of interest compared with that of the actin signal for the same evaluated RNA. Probes included chitinase (a 1.1-kb fragment of the genomic clone of the chitinase gene from pea, obtained from Dr. Lee Hadwiger at Washington State University, Pullman, WA; Chang et al., 1995), cyclin (1.5 kb cDNA of cyclin gene of parsley; Logemann et al., 1995); CDK1 (1.16-kb cDNA of cyclin dependent kinase of pea, obtained from Thomas Jacobs, University of Illinois, Champaign-Urbana), H4 histone (a 550-bp cDNA of histone 4 of parsley; Logemann et al., 1995), H3 (a 600-bp cDNA of histone 3 of parsley; Logemann et al., 1995), and, as a control, a 600-bp cDNA fragment of actin (Gunawardena and Hawes, 2002).
Statistical Analysis
Statistical analysis was done using paired data analysis with Student's t test.
Acknowledgments
This work was a portion of a Ph.D. dissertation by the first author (Department of Plant Pathology, College of Agriculture and Life Sciences, University of Arizona).
This work was supported by the U.S. Department of Agriculture (grants awarded to M.C.H.).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.104.056366.
References
- Arriola L, Niemira BA, Safir GR (1997) Border cells and arbuscular mycorrhizae in four Amaranthaceae species. Phytopathology 87: 1240–1242 [DOI] [PubMed] [Google Scholar]
- Atkinson TG, Neal JL, Larson RI (1975) Genetic control of the rhizosphere microflora of wheat. In GW Bruehl, ed, Biology and Control of Soil-borne Plant Pathogens. American Phytopathological Society, St. Paul, pp 116–122
- Ayers WA, Thornton RH (1968) Exudation of amino acids by intact and damaged roots of wheat and peas. Plant Soil 28: 193–207 [Google Scholar]
- Baluška F, Volkmann D, Barlow PW (1996) Specialized zones of development in roots: view from the cellular level. Plant Physiol 112: 3–4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bar-Yosef B (1996) Root excretions and their environmental effects: influence on availability of phosphorus. In Y Waisel, A Eshel, U Kafkafi, eds, Plant Roots: The Hidden Half. Marcel Dekker, New York, pp 581–606
- Bauer WD (1981) Infection of legumes by rhizobia. Annu Rev Plant Physiol 32: 407–449 [Google Scholar]
- Brigham LA, Woo HH, Wen F, Hawes MC (1998) Meristem-specific suppression of mitosis and a global switch in gene expression in the root cap of pea (Pisum sativum L.) by endogenous signals. Plant Physiol 118: 1223–1231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruehl GW (1986) Soilborne Plant Pathogens. Macmillan, New York
- Brunner I, Scheidegger C (1992) Ontogeny of synthesized Picea abies (L.) Karst.—Hebeloma crustuliniforme (Bull. ex St Amans) Quel ectomycorrhizas. New Phytol 120: 359–369 [Google Scholar]
- Chang CM, Horowitz D, Culley D, Hadwiger L (1995) Molecular cloning and characterization of a pea chitinase gene expression in response to wounding, fungal infection and the elicitor chitosan. Plant Mol Biol 28: 105–111 [DOI] [PubMed] [Google Scholar]
- Chi CC, Childers WR, Hanson EW (1964) Penetration and subsequent development of three Fusarium species in alfalfa and red clover. Phytopathology 54: 434–436 [Google Scholar]
- Cook RJ, Flentje NT (1967) Chlamydospore germination and germling survival of Fusarium solani f. sp. pisi in soil as affected by soil water and pea seed exudation. Phytopathology 57: 178 [Google Scholar]
- Cooper JE, Rao JR (1992) Localized changes in flavonoid biosynthesis in roots of Lotus pedunculatus after infection by Rhizobium loti. Plant Physiol 100: 444–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curl EA, Truelove B (1986) The Rhizosphere. Springer-Verlag, New York
- d'Arcy-Lameta A (1986) Study of soybean and lentil root exudates. Plant Soil 92: 113–123 [Google Scholar]
- Deacon JW (1996) Ecological implications of recognition events in the pre-infection stages of root pathogens. New Phytol 133: 135–145 [Google Scholar]
- Delserone LM, McCluskey K, Matthews DE, VanEtten HD (1999) Pisatin demethylation by fungal pathogens and non pathogens of pea: association with pisatin tolerance and virulence. Physiol Mol Plant Pathol 55: 317–326 [Google Scholar]
- deWit-Elshove A, Fuchs A (1971) The influence of the carbohydrate source on pisatin breakdown by fungi pathogenic to pea (Pisum sativum). Physiol Plant Pathol 1: 17–24 [Google Scholar]
- Dische Z (1962) General color reactions. In RL Whistler, ML Wolfrom, F Shafizadeh, eds, Methods in Carbohydrate Chemistry: Analysis and Preparation of Sugars, Vol I. Academic Press, New York, pp 478–481
- Dunn AK, Klimowicz AK, Handelsman J (2003) Use of a promoter trap to identify Bacillus cereus genes regulated by tomato seed exudate and a rhizosphere resident, Pseudomonas aureofaciens. Appl Environ Microbiol 69: 1197–1205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endre G, Kereszt A, Kevei Z, Mihacea S, Kalo P, Kiss GB (2002) A receptor kinase gene regulating symbiotic nodule development. Nature 417: 962–966 [DOI] [PubMed] [Google Scholar]
- Ferguson BJ, Mathesius U (2003) Signaling interactions during nodule development. J Plant Growth Regul 22: 47–72 [Google Scholar]
- Foster RC, Rovira AD, Cock TW (1983) Ultrastructure of the Root-Soil Interface. American Phytopathological Society, St. Paul
- Funnell DL, Matthews PS, VanEtten HD (2001) Breeding for highly fertile isolates of Nectria haematococca MPVI that are highly virulent on pea and in planta selection for virulent recombinants. Phytopathology 91: 92–101 [DOI] [PubMed] [Google Scholar]
- Funnell DL, Matthews PS, VanEtten HD (2002) Identification of new pisatin demethylase genes (PDA5 and PDA7) in Nectria haematococca and non-Mendelian segregation of pisatin demethylating activity and virulence on pea due to loss of chromosomal elements. Fungal Genet Biol 37: 121–133 [DOI] [PubMed] [Google Scholar]
- Fusconi A (1983) The development of the fungal sheath on Cistus incanus short roots. Can J Bot 61: 2546–2553 [Google Scholar]
- Garcia-Garrido JM, Ocampo JA (2002) Regulation of the plant defence response in arbuscular mycorrhizal symbiosis. J Exp Bot 53: 1377–1386 [PubMed] [Google Scholar]
- Gilbert GS, Clayton MK, Handelsman J (1996) Use of cluster and discriminant analyses to compare rhizosphere bacterial communities following biological perturbation. Microb Ecol 32: 123–147 [DOI] [PubMed] [Google Scholar]
- Goldberg NP, Hawes MC, Stanghellini ME (1989) Specific attraction to and infection of cotton root cap cells by zoospores of Pythium dissotocum. Can J Bot 67: 1760–1767 [Google Scholar]
- Graham TL (1991) Flavonoid and isoflavonoid distribution in developing soybean seedling tissues and in seed and root exudates. Plant Physiol 95: 594–603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin GJ, Hale MG, Shay FJ (1976) Nature and quantity of sloughed organic matter produced by roots of axenic peanut plants. Soil Biol Biochem 8: 29–32 [Google Scholar]
- Gunawardena U, Hawes MC (2002) Tissue specific localization of root infection by fungal pathogens: role of root border cells. Mol Plant-Microbe Interact 15: 1128–1136 [DOI] [PubMed] [Google Scholar]
- Han Y, Liu X, Benny U, Kistler HC, Van Etten HD (2001) Genes determining pathogenicity to pea are clustered on a supernumerary chromosome in the fungal plant pathogen, Nectria haematococca. Plant J 25: 305–314 [DOI] [PubMed] [Google Scholar]
- Harmsen GW, Jager G (1962) Determination of the quantity of carbon and nitrogen in the rhizosphere of young plants. Nature 195: 1119–1120 [Google Scholar]
- Harrison MJ, Dixon RA (1993) Isoflavonoid accumulation and expression of defense gene transcripts during the establishment of VAM associations in roots of Medicago truncatula. Mol Plant-Microbe Interact 6: 643–654 [Google Scholar]
- Hawes MC (1983) Sensitivity of isolated root cap cells and protoplasts to victorin. Physiol Plant Pathol 22: 65–76 [Google Scholar]
- Hawes MC, Bengough GA, Cassab G, Ponce G (2003) Root caps and rhizosphere. J Plant Growth Regul 21: 352–367 [Google Scholar]
- Hawes MC, Brigham LA, Wen F, Woo HH, Zhu Y (1998) Function of root border cells in plant health: pioneers in the rhizosphere. Annu Rev Phytopathol 36: 311–327 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Gunawardena U, Miyasaka S, Zhao X (2000) The role of root border cells in plant defense. Trends Plant Sci 5: 128–132 [DOI] [PubMed] [Google Scholar]
- Hawes MC, Wheeler H (1982) Factors affecting victorin induced cell death: temperature and plasmolysis. Physiol Plant Pathol 20: 137–144 [Google Scholar]
- Hiltner L (1904) Uber neuere Erfahrungen und Probleme auf dem Gebiet der Bodenbakteriologie und unter besonderer Berucksichtigung der Grundungung und Brache. Arb Dtsch Landwirt Ges 98: 59–78 [Google Scholar]
- Hirsch AM, Bauer WD, Bird DM (2003) Molecular signals and receptors: controlling rhizosphere interactions between plants and other organisms. Ecology 84: 858–868 [Google Scholar]
- Hirsch AM, Kapulnik Y (1998) Signal transduction pathways in mycorrhizal associations: comparisons with the Rhizobium-legume symbiosis. Fungal Genet Biol 23: 205–212 [DOI] [PubMed] [Google Scholar]
- Horan DP, Chilvers GA, Lapeyrie FF (1988) Time sequence of the infection process in eucalypt ectomycorrhizas. New Phytol 109: 451–458 [Google Scholar]
- Hsieh MC, Graham TL (2001) Partial purification and characterization of a soybean beta-glucosidase with high specific activity towards isoflavone conjugates. Phytochemistry 58: 995–1005 [DOI] [PubMed] [Google Scholar]
- Katan J (1996) Interactions of roots with soil-borne pathogens. In Y Waisel, A Eshel, U Kafkafi, eds, Plant Roots: The Hidden Half, Marcel Dekker, New York, pp 811–822
- Knee EM, Gong FC, Gao MS, Teplitski M, Jones AR, Foxworth A, Mort AJ, Bauer WD (2001) Root mucilage from pea and its utilization by rhizosphere bacteria as a sole carbon source. Mol Plant-Microbe Interact 14: 775–784 [DOI] [PubMed] [Google Scholar]
- Knudson L (1919) Viability of detached root cap cells. Am J Bot 6: 309–310 [Google Scholar]
- Kraft JM (1974) The influence of seedling exudates on the resistance of peas to Fusarium and Pythium root rot. Phytopathology 70: 981 [Google Scholar]
- Kroes GMLW, Baayen RP, Lange W (1998) Histology of root rot of flax seedlings (Linum usitatissimum) infected by Fusarium oxysporum F. sp. Lini. Eur J Plant Pathol 104: 725–736 [Google Scholar]
- Lagopodi AL, Ram AFJ, Lamers GEM, Punt PJ, Van den Hondel CA, Lugtenberg BJJ, Bloemberg GV (2002) Novel aspects of tomato root colonization and infection by Fusarium oxysporum f. sp. radicis-lycopersici revealed by confocal laser scanning microscopic analysis using the green fluorescent protein as a marker. Mol Plant-Microbe Interact 15: 172–179 [DOI] [PubMed] [Google Scholar]
- Leemhuis J, Boutillier S, Schmidt G, Meyer DK (2002) The protein kinase A inhibitor H89 acts on cell morphology by inhibiting Rho kinase. J Pharmacol Exp Ther 300: 1000–1007 [DOI] [PubMed] [Google Scholar]
- Limpens E, Bisseling T (2003) Signaling in symbiosis. Curr Opin Plant Biol 6: 343–350 [DOI] [PubMed] [Google Scholar]
- Lockwood JL (1988) Evolution of concepts associated with soilborne plant pathogens. Annu Rev Phytopathol 26: 93–121 [Google Scholar]
- Logemann E, Wu S, Schroder J, Schmeizer E, Sommssich IE, Hahlbrook K (1995) Gene activation by UV light, fungal elicitor or fungal infection in Petroselinum crispum is correlated with repression of cell cycle-related genes. Plant J 8: 865–876 [DOI] [PubMed] [Google Scholar]
- Loh J, Carlson RW, York WS, Stacey G (2002. a) Bradyoxetin, a unique chemical signal involved in symbiotic gene regulation. Proc Natl Acad Sci USA 99: 14446–14451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh J, Pierson EA, Pierson LS, Stacey G, Chatterjee A (2002. b) Quorum sensing in plant-associated bacteria. Curr Opin Plant Biol 5: 285–290 [DOI] [PubMed] [Google Scholar]
- Loria R, Lacy ML (1979) Mechanism of increased susceptibility of bleached pea seeds to seed and seeding rot. Phytopathology 69: 573 [Google Scholar]
- Maxwell CA, Phillips DA (1990) Concurrent synthesis and release of nod-gene-inducing flavonoids from alfalfa roots. Plant Physiol 93: 1552–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDougall BM, Rovira AD (1970) Sites of exudation of C-labelled compounds from wheat roots. New Phytol 69: 999–1003 [Google Scholar]
- Nagahashi G, Douds DD (2004) Isolated root caps, border cells, and mucilage from host roots stimulate hyphal branching of the arbuscular mycorrhizal fungus, Gigaspora gigantea. Mycol Res 108: 1079–1088 [DOI] [PubMed] [Google Scholar]
- Nair MG, Safir GR, Siqueira JO (1991) Isolation and identification of VAM stimulatory compounds from clover (Trifolium repens) roots. Appl Environ Microbiol 57: 434–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson EB (1991) Exudate molecules initiating fungal responses to seeds and roots. In DL Keister, PB Cregan, eds, The Rhizosphere and Plant Growth. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 197–209
- Nelson EB (2004) Microbial dynamics and interactions in the spermosphere. Annu Rev Phytopathol 42: 271–309 [DOI] [PubMed] [Google Scholar]
- Niemira BA, Safir GR, Hawes MC (1996) Arbuscular mycorrhizal colonization and border cell production: a possible correlation. Phytopathology 86: 563–565 [Google Scholar]
- O'Connell KP, Goodman RM, Handelsman J (1996) Engineering the rhizosphere: expressing a bias. Trends Biotechnol 14: 83–88 [Google Scholar]
- Parkinson D (1955) Liberation of amino acids by oat seedlings. Nature 176: 35–36 [Google Scholar]
- Peters NK, Long SR (1988) Alfalfa root exudates and compounds which promote or inhibit induction of Rhizobium meliloti nodulation genes. Plant Physiol 88: 396–400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips DA, Fox TC, King MD, Bhuvaneswari TV, Teuber LR (2004) Microbial products trigger amino acid exudation from plant roots. Plant Physiol 136: 2887–2894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Recourt K, Verkerke M, Schripsema J, vanBrussel AAN, Lugtenberg BJJ, Kijne JW (1992) Major flavonoids in uninoculated and inoculated roots of Vicia sativa subsp. nigra are four conjugates of the nodulation gene-inhibitor kaempferol. Plant Mol Biol 18: 505–513 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Galvez E, Mendgen K (1995) The infection process of Fusarium oxysporum in cotton root tips. Protoplasma 189: 61–72 [DOI] [PubMed] [Google Scholar]
- Roth M (1971) Fluorescence reaction for amino acids. Anal Chem 43: 880–882 [DOI] [PubMed] [Google Scholar]
- Rovira AD (1956) Plant root excretions in relation to the rhizosphere effect. 1. The nature of root exudate from pea and oats. Plant Soil 7: 178–194 [Google Scholar]
- Rovira AD (1959) Plant root excretions in relation to the rhizosphere effect. IV. Influence of plant species, age of plant, light, temperature and calcium nutrition of exudation. Plant Soil 11: 53–64 [Google Scholar]
- Rovira AD (1991) Rhizosphere research—85 years of progress and frustration. In DL Keister, PB Cregan, eds, The Rhizosphere and Plant Growth. Kluwer Academic Publishers, Dordrecht, The Netherlands, pp 3–14
- Ruan Y, Kotraiah V, Straney DC (1995) Flavonoids stimulate spore germination in Fusarium solani pathogenic on legumes in a manner sensitive to inhibitors of cAMP-dependent protein kinase. Mol Plant-Microbe Interact 8: 929–938 [Google Scholar]
- Schroth MN, Tousson TA, Snyder WC (1963) Effect of certain constituents of bean exudate on germination of chlamydospores of Fusarium solani f. sp. phaseoli in soil. Phytopathology 53: 809–812 [Google Scholar]
- Sherwood R (1987) Papilla formation in corn root cap cells and leaves inoculated with Colletotrichum graminicola. Phytopathology 77: 930–934 [Google Scholar]
- Short GE, Lacy ML (1974) Germination of Fusarium solani f. sp. pisi chlamydospores in the spermosphere of pea. Phytopathology 64: 558–562 [Google Scholar]
- Short GE, Lacy ML (1976) Carbohydrate exudation from pea seeds: effect of cultivar, seed age, seed color and temperature [and relation to fungal rots]. Phytopathology 66: 182–187 [Google Scholar]
- Smith KP, Handelsman J, Goodman RM (1999) Genetic basis in plants for interactions with disease-suppressive bacteria. Proc Natl Acad Sci USA 96: 4786–4790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sommssich IE, Hahlbrock K (1998) Pathogen defense in plants—a paradigm of biological complexity. Trends Plant Sci 3: 86–90 [Google Scholar]
- Stahl DJ, Theuerkauf A, Heitefuss R, Schafer W (1994) Cutinase of Nectria haematococca (Fusarium solani F. sp. pisi) is not required for fungal virulence or organ specificity on pea. Mol Plant-Microbe Interact 7: 713–725 [Google Scholar]
- Steele HL, Werner D, Cooper JE (1999) Flavonoids in seed and root exudates of Lotus pedunculatus and their biotransformation by Mesorhizobium loti. Physiol Plant 107: 251–258 [Google Scholar]
- Stevens RB (1974) Mycology Guidebook. University of Washington Press, Seattle
- Straney DC, VanEtten HD (1994) Characterization of the PDA1 promoter of Nectria haematococca and identification of a region that binds a pisatin-responsive DNA binding factor. Mol Plant-Microbe Interact 7: 256–266 [DOI] [PubMed] [Google Scholar]
- Syin C, Parzy D, Traincard F, Boccaccio I, Joshi MB, Lin DT, Yang X, Assemat K, Doerig C, Langsley G (2001) The H89 cAMP-dependent protein kinase inhibitor blocks Plasmodium falciparum development in infected erythrocytes. Eur J Biochem 268: 4842–4849 [DOI] [PubMed] [Google Scholar]
- Teplitski M, Robinson JB, Bauer WD (2000) Plants secrete substances that mimic bacterial N-acyl homoserine lactone signal activities and affect population density-dependent behaviors in associated bacteria. Mol Plant-Microbe Interact 13: 637–648 [DOI] [PubMed] [Google Scholar]
- Tsai SM, Phillips DA (1991) Flavonoids released naturally from alfalfa promote development of symbiotic Glomus spores in vitro. Appl Environ Microbiol 57: 1485–1488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanuo MK, Hassanali A, Hooper AM, Khan Z, Kaberia F, Pickett JA, Wadhams LJ (2003) Isoflavanones from the allelopathic root exudate of Desmodium uncinatum. Phytochemistry 64: 265–273 [DOI] [PubMed] [Google Scholar]
- Turlier M, Eparvier A, Alabouvette C (1994) Early dynamic interactions between Fusarium oxysporum f. sp. lini and the roots of Linum usitatissimum as revealed by transgenic GUS-marked hyphae. Can J Bot 72: 1605–1612 [Google Scholar]
- Van Brussel AAN, Recourt K, Pees E, Splink HP, Tak T (1990) A biovar-specific signal of Rhizobium leguminosarum bv. viciae induces increased nodulation gene inducing activity in root exudate of Vicia sativa subsp. nigra. J Bacteriol 172: 5394–5401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanEgeraat AWSM (1975) The possible role of homoserine in the development of Rhizobium leguminosarum in the rhizosphere of pea seedlings. Plant Soil 42: 381–386 [Google Scholar]
- Van Etten HD, Matthews PS, Tegtmeier KJ, Dietert MF, Stein JI (1980) The association of pisatin tolerance and demethylation with virulence on pea in Nectria haematococca. Physiol Plant Pathol 16: 257–268 [Google Scholar]
- Vermeer J, McCully ME (1982) The rhizosphere in Zea: new insight into its structure and development. Planta 156: 45–61 [DOI] [PubMed] [Google Scholar]
- Wassman CC, Van Etten HD (1996) Transformation-mediated chromosome loss and disruption of a gene for pisatin demethylase decrease the virulence of Nectria haematococca on pea. Mol Plant-Microbe Interact 9: 793–803 [Google Scholar]
- Wojtaszek P, Stobiecki M, Gulewicz K (1993) Role of nitrogen and plant growth regulators in the exudation and accumulation of isoflavonoids by roots of intact white lupin (Lupinus albus L.) plants. J Plant Physiol 142: 689–694 [Google Scholar]
- Woo HH, Hirsch AM, Hawes MC (2004) Altered susceptibility to infection by Sinorhizobium meliloti and Nectria haematococca in alfalfa roots with altered cell cycle. Plant Cell Rep 22: 967–973 [DOI] [PubMed] [Google Scholar]
- Zhu Y, Pierson LS III, Hawes MC (1997) Induction of microbial genes for pathogenesis and symbiosis by chemicals from root border cells. Plant Physiol 115: 1691–1698 [DOI] [PMC free article] [PubMed] [Google Scholar]