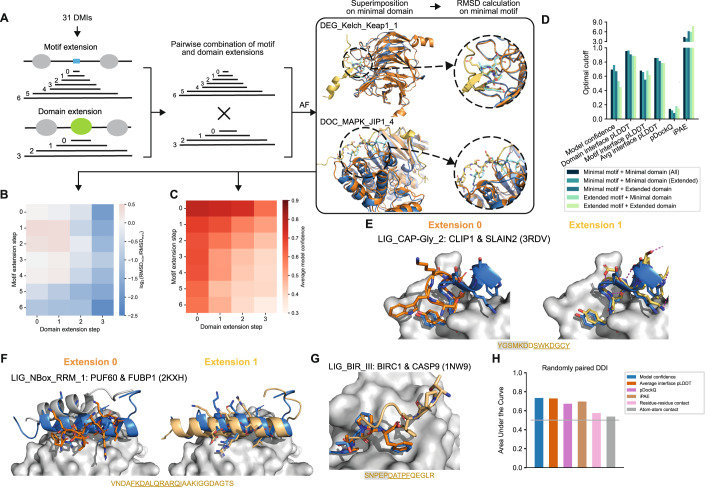
Figure 2. Effect of protein fragment extensions on the accuracy of AF predictions.
(A) Workflow established to assess changes in AF performance upon protein fragment extension. Blue and cyan indicate the domain and motif in the native structure, respectively. Orange and yellow indicate the domain and motif in the modeled structure, respectively. (B) Heatmap showing the fold change in motif RMSD before and after extension where positive values indicate improved predictions from extension and negative values indicate worse prediction outcomes upon extension. (C) Heatmap of the average model confidence for combinations of different motif and domain sequence extensions. (D) Optimal cutoffs derived for different metrics from ROC analysis benchmarking AF different motif and domain extensions from the reference dataset used in A and random pairings of domain and motif sequences. pLDDT-related metrics were divided by 100 for visualization purposes. (E, F) Superimposition of the structural model of the minimal (left, orange) or extended (right, yellow) motif sequence with the solved structure (motif in blue) for two different motif classes as indicated on the top of each panel. The motif sequence from the solved structure is indicated at the bottom. Motif residues are underlined, motif residues not resolved in the structure have a gray background. Sticks indicate the motif residues, domain surfaces are shown in gray based on experimental structures. (G) Superimposition of the structural model of the minimal (orange) and extended (yellow) motif sequence with the solved structure (motif in blue) for a motif instance from the motif class LIG_BIR_III. Motif sequence indicated as in (E). (H) Area under the Receiver Operating Characteristics Curve (AUROC) for different metrics using the DDI benchmark dataset as positive reference and randomly shuffled domain-domain pairs as random reference. Gray horizontal line indicates the AUROC of a random predictor.