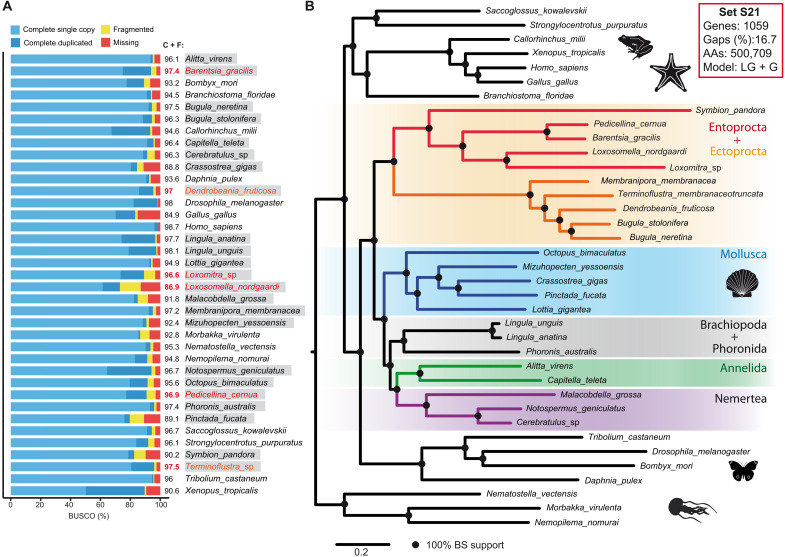
Fig. 2. Quality of the datasets used and the ML tree based on 1059 protein markers.
(A) Quality assessment of the datasets used for phylogeny reconstruction by BUSCO. Completeness (C + F, complete + fragmented) of all newly generated proteins sets, except Loxosomella, exceeds 96.6%, which corresponds to the quality range typical for genome projects. Lophotrochozoan species are highlighted with gray background, and six newly sequenced species are shown in red. (B) ML tree based on concatenation of 1059 protein markers present in all 37 species. LG + G substitution model, 500,709 distinct alignment patterns, and all branches have maximum BS. AAs, amino acids.