Figure 2.
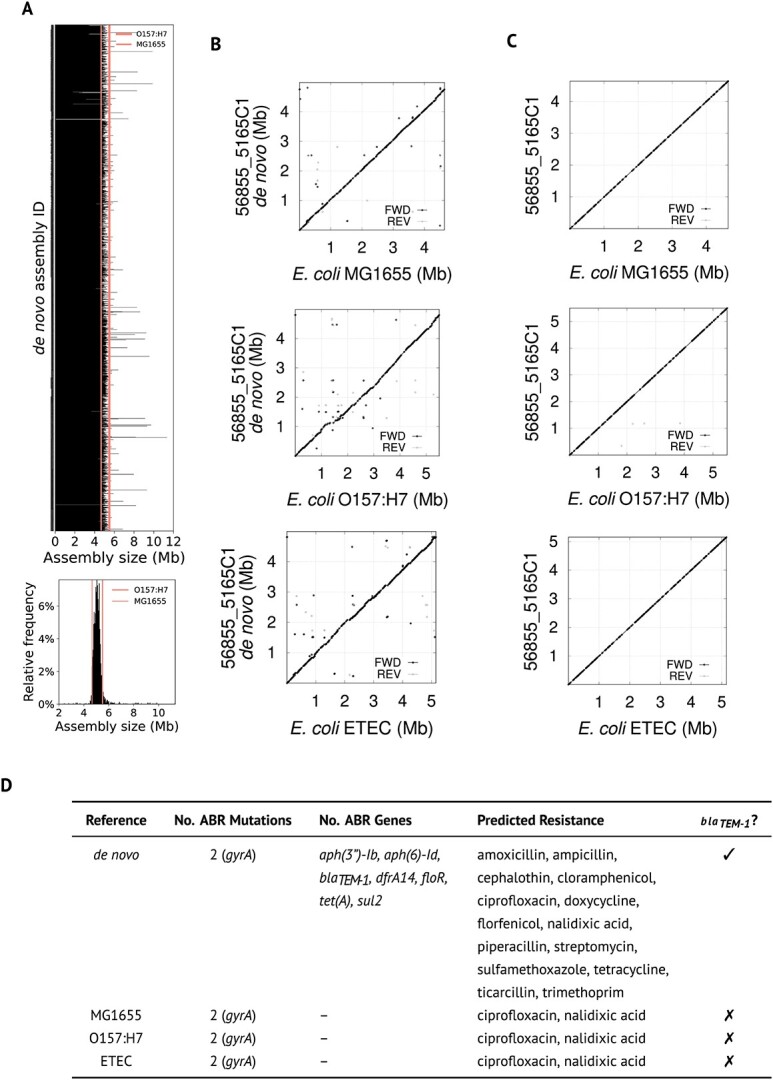
Variability of within species genome size is not captured by the canonical use of reference genomes. (A) Histogram (top) and distribution (bottom) of genome sizes across 3562 farm and clinical E. coli isolates. The canonical size of the reference wild-type (K-12 substr. MG1655, genome GCF_000005845.2 in the NCBI) and Shiga toxin-producing strain (0157:H7 str. Sakai, genome GCA_000008865.2 in the NCBI) are marked at ~4.64 Mb and 5.59 Mb by vertical lines. (B) and (C) Dotplots of one representative isolate to visualize the genome–genome sequence alignment between de novo (B) or reference-mapped (C) assemblies and three different known genomes: MG1655, O157:H7, and enterotoxigenic E. coli (ETEC). Sequences present in these references, but not in out isolate, are noted by a horizontal gap in the dotplot, whereas the converse is noted by a vertical gap. Note that in C) the reported assembly size of our isolate, in the y-axis, varies depending on the choice of reference—size is constant in those genomes assembled de novo. (D) Exemplar report from ResFinder when used against the isolate 56855_5165C1 from B to C assembled de novo, and mapped to different references. This tool reports documented ABR mutations and genes, as well as predicting resistance to certain antibiotics as reported by the literature. Note that, for the de novo assembly, ResFinder reports resistance to ampicillin and amoxicillin. The number of copies of blaTEM-1 reported by Hound is 2–3, thus, the isolate would also be resistant to amoxicillin/clavulanate [28] which is missed by ResFinder. The full report can be found in the supplementary data.
