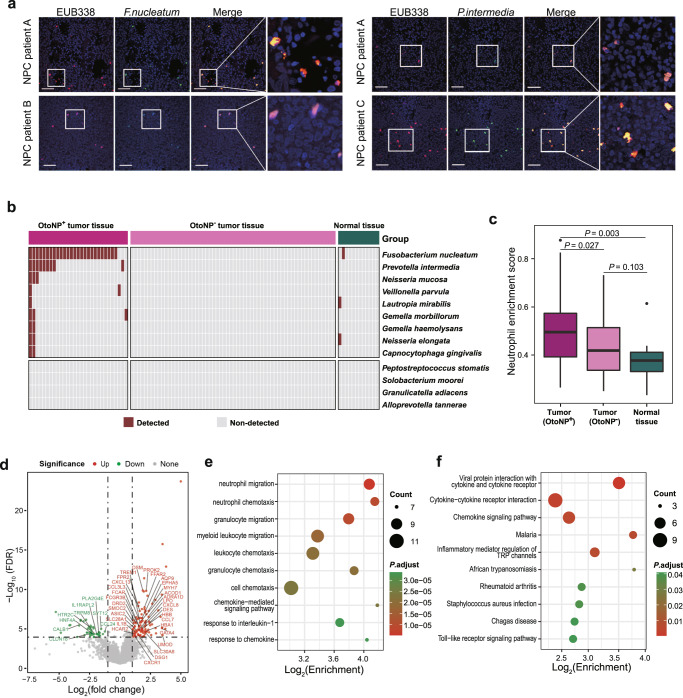
Fig. 4. Oral translocated microbes infiltrated the tumor and affected the tumor microenvironment.
a Representative images of the presence of Fusobacterium nucleatum (left) and Prevotella intermedia (right) in tumor tissues of NPC patients. EUB338 (red) was a Cy3-conjugated universal bacterial oligonucleotide probe. F. nucleatum and P. intermedia were FITC-conjugated oligonucleotide probes (green). High-magnification images of the boxed area are shown on the right. Scale bars, 50 μm. Three and four patients were identified for F. nucleatum and P. intermedia with similar results. b The detection status of “NPCOtoNP” microbes in meta-transcriptomic sequencing data of nasopharyngeal tissues (N = 29 for OtoNP+ tumors, N = 60 for OtoNP- tumors and N = 12 for normal tissues). c The boxplot showing the enrichment score for neutrophil among OtoNP+ tumors (N = 29), OtoNP- tumors (N = 60) and normal tissues (N = 12). P values were determined by t test (two-sided). Boxplots were presented with the median marked by thick black line, the interquartile range marked by the bar, the range by the thin line and outliners by the black dots (d) The volcano plot showing the differentially expressed genes between OtoNP+ (N = 29) and OtoNP- (N = 60) tumor tissues (edgeR, two-sided, FDR-q < 0.05 & |log2FC| > 1). Genes involved in top 10 significant GO and KEGG pathways were labeled in the plot. The results of pathways enrichment analysis of differential genes based on GO (e) and KEGG pathways (f). “NPCOtoNP” microbes, the species that NPC-enriched and high-translocation-enriched; All pathways were with the FDR-adjusted P < 0.05 (two-sided). OtoNP+ tumor, the tumor with “NPCOtoNP” microbes; OtoNP− tumor, the tumor without “NPCOtoNP” microbes, FISH fluorescence in situ hybridization probes, ssGSEA single sample Gene Set Enrichment Analysis, GO Gene Ontology, KEGG Kyoto Encyclopedia of Genes and Genomes, FDR false discovery rate. Source data are provided as a Source Data file.