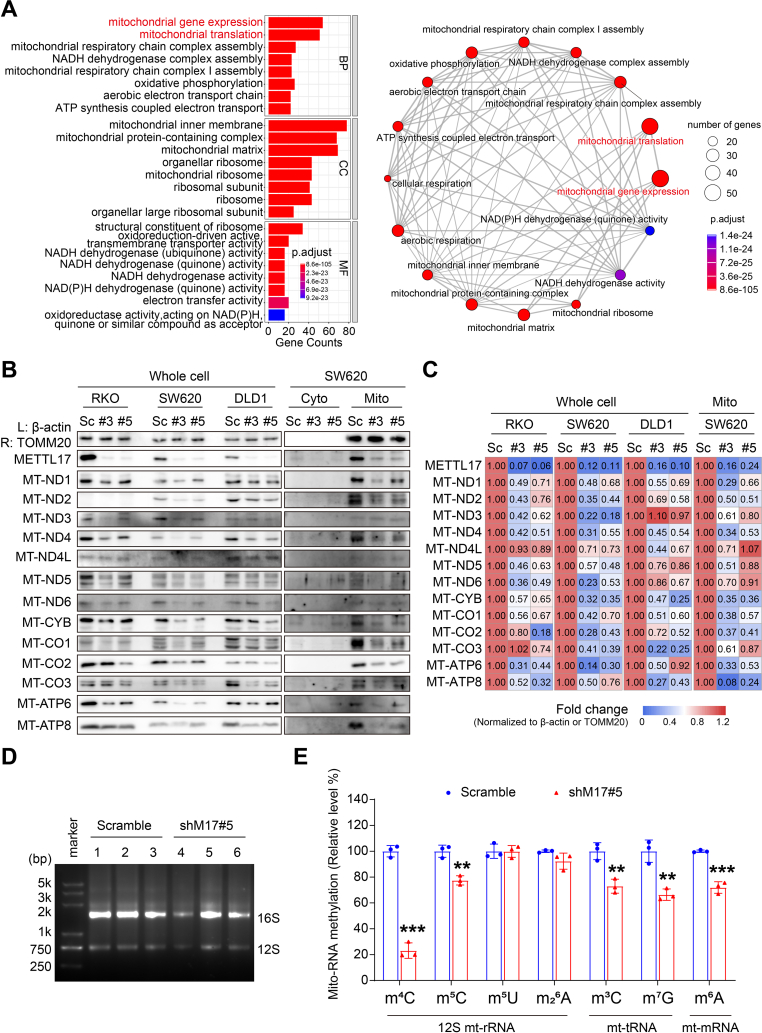
Fig. 5.
METTL17 is required for global mitochondrial translation in mitochondrial RNA methylation manner in CRC.
A. GO enrichment analysis revealed that the top 100 METTL17 DepMap co-dependent genes predominantly associated with mitochondria, influencing the regulation of mitochondrial gene expression and mitochondrial translation. The bar plot illustrated enrichments in multiple mitochondria-related gene sets (left), while the visualized network highlighted the close connections among these gene sets (right). BP, biological process. CC, cellular component. MF, molecular function.
B and C. Knockdown of METTL17 down-regulated the protein expression of mitochondrial coding genes in CRC cells detected by Western blot (B). Semi-quantitative analysis of the relative protein levels was performed and visualized in the heatmap of fold change, normalized to Scramble (C).
D and E. The quality of mitochondrial RNA from RKO cells was identified by agarose gel assay (D). HPLC-MS analysis of mitochondrial RNA showed that knockdown of METTL17 significantly reduced m4C, m5C, m3C, m7G, and m6A levels within mitochondrial RNA methylations in RKO cells (E). Relative levels were quantified by comparing to scramble, n = 3 per group.
Data are shown as mean ± SD. **p < 0.01, ***p < 0.001 compared to Scramble group, based on a two-sided Student's t-test.