FIGURE 6.
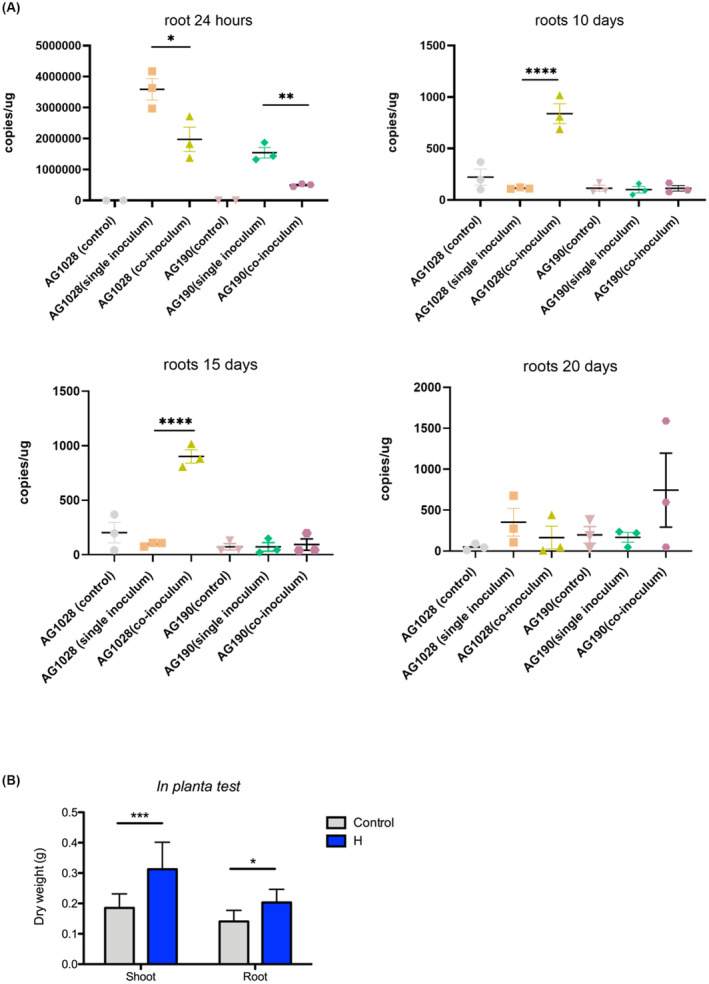
(A) qPCR analysis for the presence of Pseudomonas fulva AG1028 and Bacillus megaterium AG190 in rice roots. Bacterial DNA copies/μg of root grown in Hoagland's solution with three different treatments with AG190 (B. megaterium) and AG1028 (P. fulva). Inoculated 7‐days young rice plant roots samples collected 24 h, 10, 15, and 20 days. Control means non‐inoculated; single inoculum means just AG190 (B. megaterium) or AG1028 (P. fulva) inoculated, and co‐inoculum means AG190 (B. megaterium) and AG1028 (P. fulva) co‐inoculated. Three biological replicates were used in each group. Two‐way analysis of variance (ANOVA) with Tukey's post hoc multiple comparison test was used for multigroup comparison analysis. All data are presented as means ± SEM. GraphPad Prism 8.2 (GraphPad Software, Inc.) was used to perform all statistical analyses. *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 as indicated. The error bars indicate SDs. (B) Plant growth promotion assays performed on rice of the 2‐strain bacterial consortia (P. fulva AG1028 and B. megaterium AG190) under normal growth conditions at 20 days post inoculation. See text for all details.
