Figure 4.
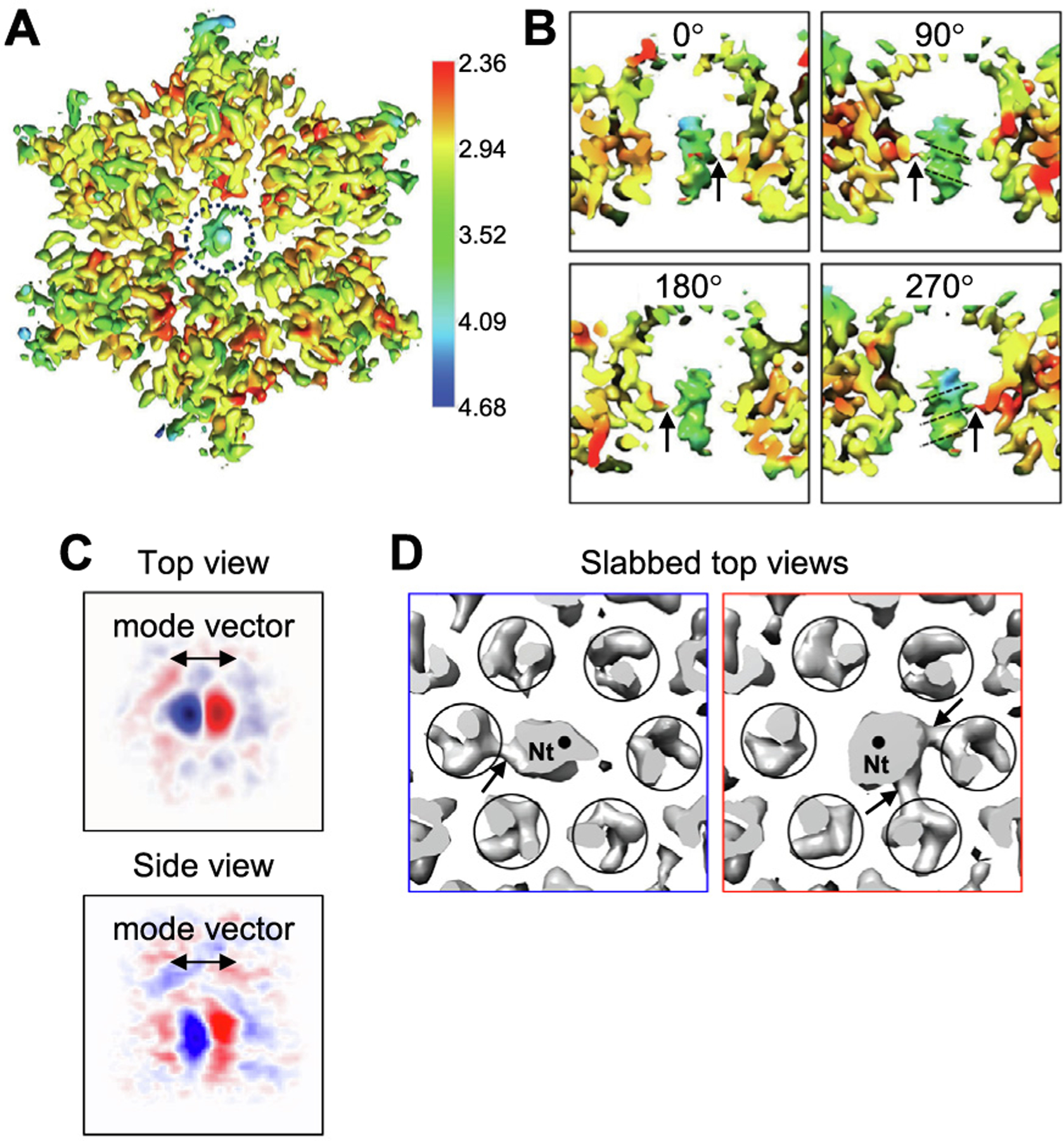
CryoEM structure of the CA hexamer bound to Nt. (A) Top view of the consensus map refined with C1 symmetry, colored according to local resolution. The peptide in the central channel is encircled. (B) Orthogonal side views of the peptide density. Arrows indicate apparent contacts between the peptide and Arg18. Dashed lines indicate evenly spaced ridges that we interpret to be consistent with a predominant helical configuration. (C) Orthogonal views of central slices through the major 3D variability component for the consensus map, as determined by 3DVA analysis. Positive (red) and negative (blue) values indicate different positions of the peptide relative to the central hexamer channel. (D) Reconstructions of particle subsets, without further alignment, revealing two peptide orientations determined by 3DVA. Circles indicate densities for CA helix 1 surrounding the central channel. Black dots indicate the central axis of the hexamer channel. Arrows indicate clearly defined contacts between the peptide and Arg18.
