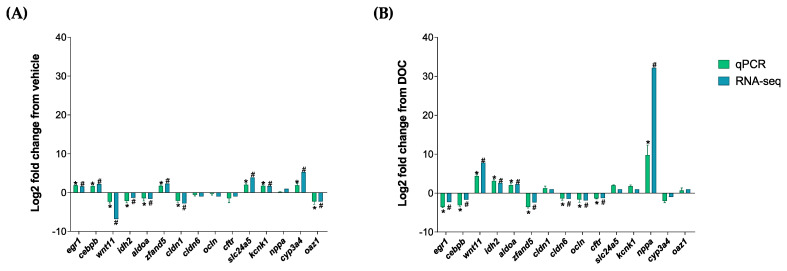
Figure 5.
RNA-seq analysis validation via a real-time PCR of the DETs. The following DETs were selected for the real time-PCR validation of the RNA-seq process: egr1, cebpb, wnt11, idh2, aldoa, zfand5, cldn1, cldn6, ocln, cftr, slc24a5, kcnk1, nppa, cyp3a4, and oaz1. (A) Validation results between the vehicle vs. DOC and (B) the DOC vs. eplerenone + DOC. For the RNA-seq, “#” in blue indicates a padj of <0.05 and an absolute Log2FC of >1. For the real-time PCR (n = 3), the relative expression was normalized against actβ and fau, and the “*” in green indicates the significant differences in the fold change in the vehicle or DOC groups (mean ± SEM, n = 3, p < 0.05). The genes had the following abbreviations: egr1 (early growth response 1); cebpb (CCAAT enhancer binding protein beta); wnt11 (Wnt family member 11); idh2 (isocitrate dehydrogenase (NADP(+)) 2); aldoa (aldolase, fructose-bisphosphate A); zfand5 (zinc finger AN1-type containing 5); cldn1 (claudin 1); cldn6 (claudin 6); ocln (occludin); cftr (cystic fibrosis transmembrane conductance regulator); slc24a5 (solute carrier family 24 member 5); kcnk1 (potassium two-pore domain channel subfamily K member 1); nppa (natriuretic peptide A); cyp3a4 (cytochrome P450 family 3 subfamily A member 4); oaz1 (ornithine decarboxylase antizyme 1); fau (40S ribosomal protein S30); and actβ (beta-actin).