FIG 2.
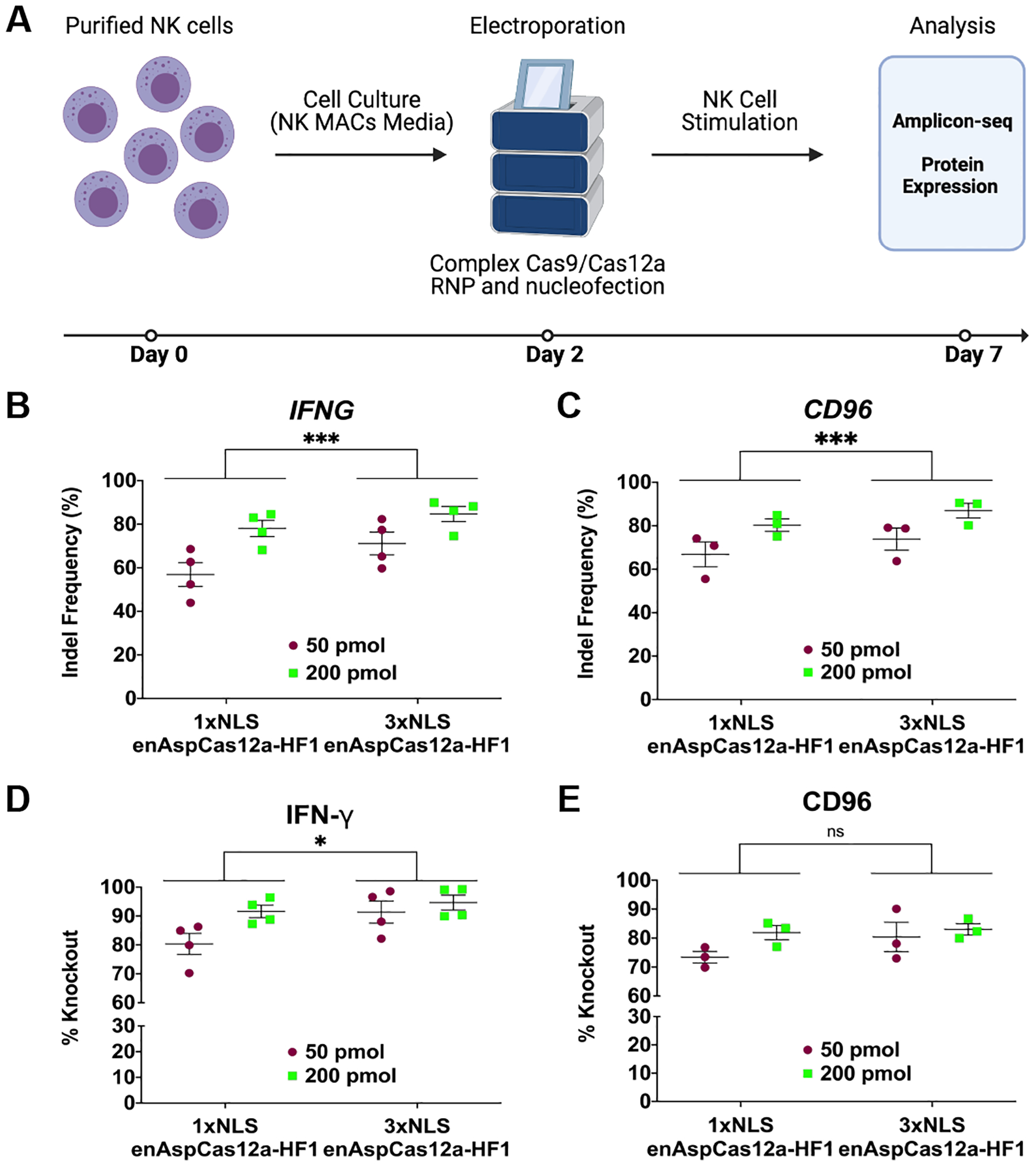
Robust gene editing in NK cells by enAspCas12a. (A) Schematic representation of experimental workflow for NK editing and phenotypic analysis. (B) Quantification of nuclease activity at IFNG locus (5 days post-nucleofection) based on Illumina sequencing. (C) Quantification of nuclease activity at CD96 locus (5 days post-nucleofection) based on Illumina sequencing. (D) Quantification of %Knockout of IFN-γ cytokine in stimulated NK cells. Cells are stimulated for 3 hours (PMA+ionomycin) or overnight (IL-12+IL-15+IL-18 cocktail). The fraction of cells positive for protein expression was determined by flow cytometry. (E) Quantification of %Knockout of CD96 protein in stimulated NK cells. Cells are stimulated for 3 hours (PMA+ionomycin) or overnight (IL-12+IL-15+IL-18 cocktail). The fraction of cells positive for protein expression was determined by flow cytometry. Results were obtained from four (IFNG) and three (CD96) independent experiments and presented as mean ± SEM. *P < 0.05, **P < 0.01, ***P < 0.001 by two-way ANOVA with Tukey’s multiple comparisons test between 1xNLS and 3xNLS enAspCas12a.
