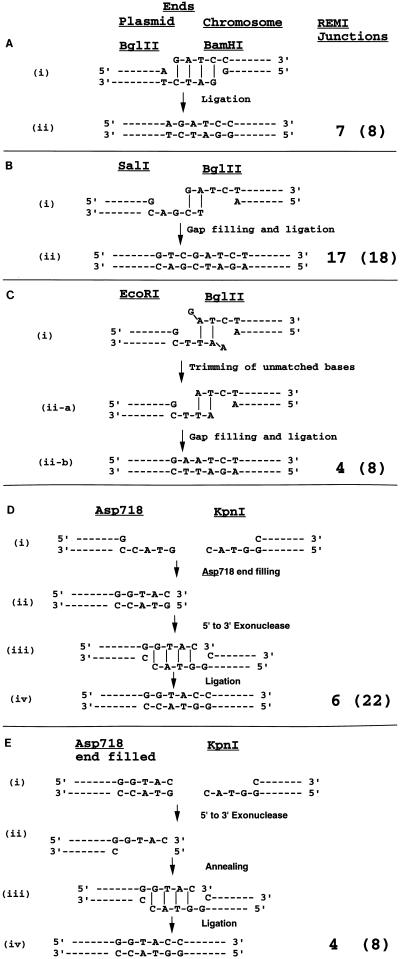
FIG. 3.
Model for end joining during REMI. The number of REMI junctions that showed the structures depicted are shown in the “REMI junctions” column, and the total number of events analyzed for the particular combination is shown in parentheses. (A) BH events. Plasmid pM151 was digested with BglII and contains 5′GATC PSS ends. BamHI is added to the transformation mixture and should create the same GATC 5′ PSS ends (i). As the ends are compatible, they can anneal by microhomology. Ligation of nicks completes the integration process (ii). (B) SB events. Plasmid pM150 was digested with SalI and contains 5′TCGA PSS ends. The ends produced in vivo by BglII (GATC) have the terminal 2 bp compatible with the SalI ends (i). All the junction sequences are GTCGATCT sequences, indicating that integration took place through annealing of the two terminal bases, gap filling, and ligation of nicks (ii). (C) RB events. Plasmid pM150 was digested with EcoRI and contains 5′AATT PSS ends. The 5′ PSS ends produced in vivo by BglII (GATC) have the central 2 bp compatible with the EcoRI ends. Before or after annealing of the central two bases, the terminal unmatched bases are cleaved off, leaving a gap on both sides which is filled by DNA polymerase and ligated to seal the gap. (D) AK events. Plasmid pM150 was digested with Asp718 and contains 5′GTAC PSS ends. The 3′ PSS ends (GTAC) were produced in vivo by KpnI. Since the integration events recreated the KpnI sites, we proposed that filling of the Asp718 site by polymerase followed by 5′-3′ exonuclease digestion would create a 3′GTAC PSS end that was used as a substrate for KpnI-mediated integrations. (E) In vitro filling of an Asp718 site creates a blunt end that also works for REMI with KpnI, suggesting that the blunt-ended substrate is converted to a 3′GTAC PSS sequence by 5′-3′ exonuclease. Event types are defined in the legend to Fig. 2.