Figure 4: Conserved residues in LysinB_MF.
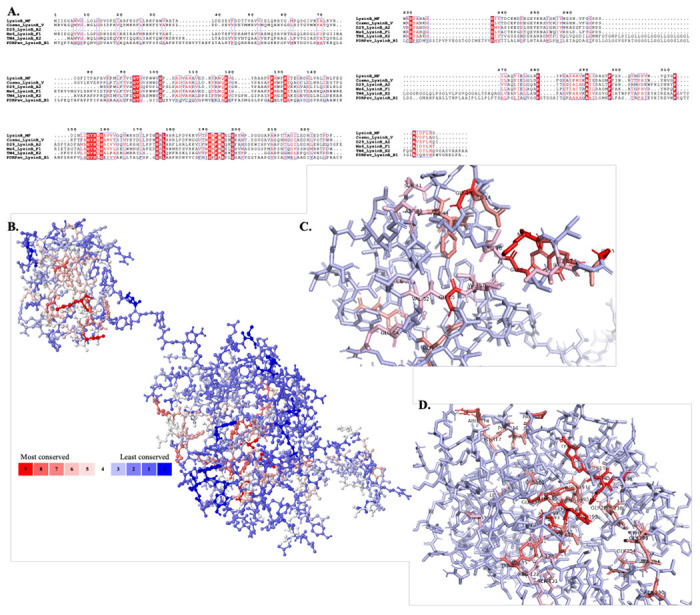
A) Multiple sequence alignment of LysinB enzymes from various clusters demonstrate key residues in PGBD and EAD. The EAD displays the conserved active-site residues Ser-Asp-His and the GXP motif. B) Analysis using POLYVIEW-MM identified and mapped conserved residues in the tertiary structure of LysinB_MF. A scale of 0-9 is used to quantify the conservation index, where 0 indicates the least conserved and nine is the most. Exposed residues in the structure of LysinB_MF are found to be least conserved among all others, while most of the highly conserved residues are found buried. C) Bird’s eye view of PGBD displays conserved amino acids. Ser41, Tyr43 Phe44, Asp45, Val52, Glu54, Gln56, Gly66 and Ile67 are a few highly conserved residues in this domain found both in sequence alignment and in structural analysis. D) Bird’s eye view of EAD displays conserved amino acids. The active site Ser156, Asp236 & His305 and Gly191 & Pro193 from the GXP motif were highly conserved in LysinBs.
