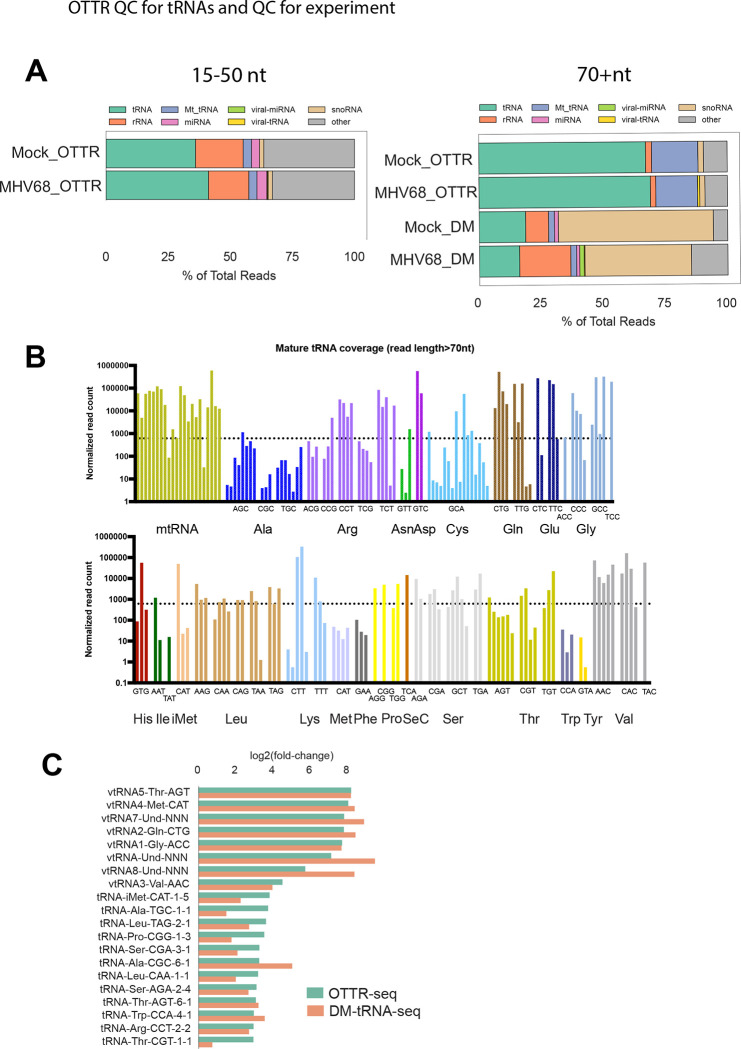
Figure 2. OTTR-seq robustly captures host tRNAs.
(A) Distribution of reads mapping to various small RNA classes using OTTR-seq (OTTR) or DM-tRNA-seq (DM) prepared from mock and MHV68 infected MC57G mouse fibroblasts at an MOI=5 for 24 hours. (B) Normalized read counts for host tRNA isodecoders detected in mock-infected libraries using OTTR-seq. The dotted line marks the median normalized read count for all tRNA isodecoders as reference. (C) Log2(fold-change) values between MHV68 infected (MR) versus Mock infection from OTTR-seq (green) or DM-tRNA-seq (orange) are consistent for viral-tRNAs (“vtRNA” prefix) and host pre-tRNAs.