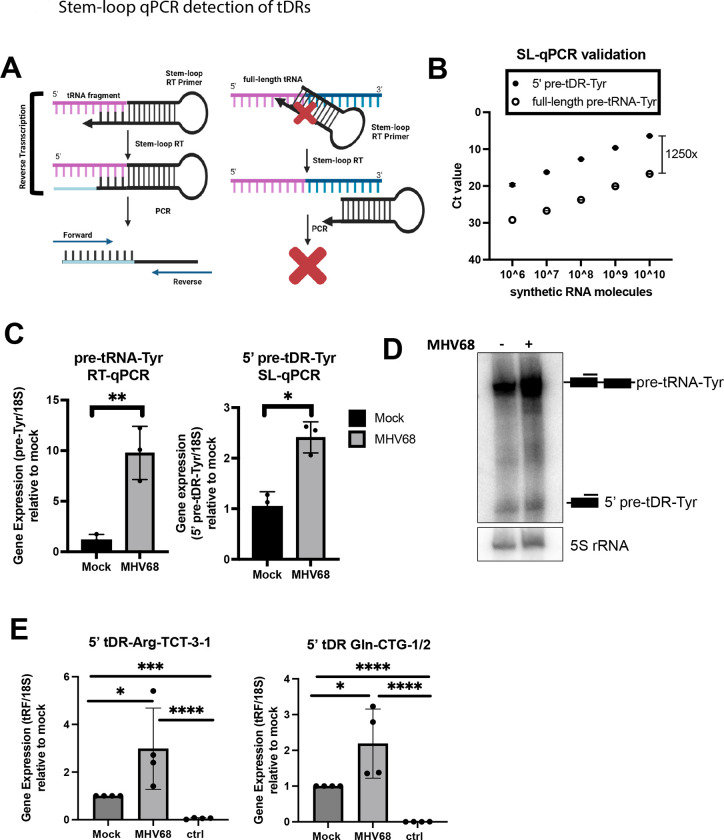
Figure 6. Stem-loop qPCR detection of tDRs.
(A) Schematic illustrating the specificity of the stem-loop (SL) RT primer for tDR vs. full-length parental tRNA. (B) Synthetic full-length pre-tRNA-Tyr or 5’ pre-tDR-Tyr were used in known quantities for SL-qPCR using Tyr forward and SL reverse primers. Raw Ct values are shown. (C) Standard- and SL-qPCR was performed using total RNA from mock or MHV68 infected NIH 3T3s to detect the full-length pre-tRNA-Tyr or 5’ pre-tDR-Tyr, respectively. Data depicts means +/− SD from three independent experiments relative to the mock sample, with p-values calculated using raw DCt values and paired t-test. (D) Northern blot using 5’ exon Tyr probes was performed using RNA samples as described in (C). (E) SL-qPCR was performed from four independent experiments to detect 5’ tDR-Arg-TCT and - Gln-CTG as in (C). The sample labeled “ctrl” was reverse transcribed with a SL RT primer for an unrelated tDR species. tDRnamer21 nomenclature for amplified tDRs is reported in Materials and Methods. ns = p>0.05; * = p ≤ 0.05; ** = p ≤ 0.01; *** = p ≤ 0.001