Abstract
Background
Retreatment with anti-EGFR monoclonal antibodies is a promising strategy in patients with RAS/BRAF wild-type (wt) metastatic colorectal cancer (mCRC) who achieved benefit from previous anti-EGFR exposure upon exclusion of mutations in RAS/BRAF genes according to circulating tumor DNA (ctDNA) analysis by means of liquid biopsy (LB). This treatment approach is now being investigated in the randomized phase II trial PARERE (NCT04787341). We here present preliminary findings of molecular screening.
Methods
Patients with RAS/BRAFV600E wt mCRC according to tissue genotyping who benefited from previous anti-EGFR-based treatment (fluoropyrimidines, oxaliplatin, irinotecan, and antiangiogenics) and then experienced disease progression to EGFR targeting were eligible for screening in the PARERE trial. The next-generation sequencing (NGS) panel Oncomine™ was employed for ctDNA testing.
Results
A total of 218 patients underwent LB, and ctDNA sequencing was successful in 201 of them (92%). RAS/BRAFV600E mutations were found in 68 (34%) patients and were mainly subclonal (median variant allele fraction [VAF] for KRAS, NRAS, and BRAF mutant clones: 0.52%, 0.62%, and 0.12%, respectively; p = 0.01), with KRASQ61H being the most frequently detected (31%). Anti-EGFR-free intervals did not predict ctDNA molecular status (p = 0.12). Among the 133 patients with RAS/BRAFV600E wt tumors according to LB, 40 (30%) harbored a mutation in at least another gene potentially implied in anti-EGFR resistance, mainly with subclonal expression (median VAF, 0.56%). In detail, alterations in PIK3CA, FBXW7, GNAS, MAP2K, ERBB2, BRAF (class I and II non-BRAFV600E), SMAD, EGFR, AKT1, and CTNNB1 occurred in 13%, 8%, 7%, 3%, 2%, 2%, 1%, 1%, 1%, and 1% cases, respectively. Co-mutations were detected in 13 (33%) out of 40 patients.
Conclusions
This is the largest prospective cohort of mCRC patients screened with LB for anti-EGFR retreatment in a randomized study. ctDNA genotyping reveals that at least one out of three patients candidate for retreatment should be excluded from this therapy, and other potential drivers of anti-EGFR resistance are found in approximately one out of three patients with RAS/BRAFV600E wt ctDNA.
Keywords: mCRC, anti-EGFR, retreatment, ctDNA, liquid biopsy, next generation sequencing
1. Introduction
Retreatment with monoclonal antibodies targeting the anti-epidermal growth factor receptors (anti-EGFRs) is a promising approach for RAS/BRAF wild-type (wt) metastatic colorectal cancer (mCRC) patients who developed acquired resistance to previous anti-EGFR exposure (1–6). However, the overall response rates (ORRs) observed in initial single-arm phase II trials range between 0% and 21%, suggesting the need for a more accurate patient selection (2–4, 6).
To this end, circulating tumor DNA (ctDNA) sequencing in liquid biopsy (LB) is regarded as the preferred approach to fine-tune the identification of patients candidate for anti-EGFR retreatment, as reported by post-hoc translational analyses of single-arm phase II studies, showing no benefit in patients with RAS/BRAF mutant ctDNA, and the recent prospective ctDNA-guided CHRONOS trial, reporting an ORR of 30% in patients with RAS/BRAF wt ctDNA (1–3, 6). Similarly, a multi-arm non-comparative phase 2 trial by Parseghian et al. showed an ORR of 20% to anti-EGFR retreatment in patients with no acquired mutations in RAS/BRAF/MAP2K/EGFR, and no responses to anti-EGFR retreatment in patients harboring one of these mutations in their ctDNA (4). Conversely, the evaluation of clinical biomarkers as surrogates of RAS/BRAF mutational status led to conflicting results (7), thus endorsing the prospective adoption of molecular screening with LB in recent studies on anti-EGFR retreatment and in the real-world setting where anti-EGFR retreatment was proven feasible and active (ORR, 25%) in patients with no RAS or BRAF mutations in their ctDNA (5, 8, 9).
These data consistently suggest that while up to one-third of patients may benefit from anti-EGFR retreatment in the case of RAS/BRAF wt ctDNA, approximately two-thirds of them still do not respond to anti-EGFR re-exposure, including one-third of rapid progressors (1, 7). Therefore, RAS and BRAF genotyping alone does not allow to catch the wide complexity of escape mechanisms to EGFR blockade, so the adoption of broader genomic panels (i.e., next-generation sequencing [NGS] assays) may further improve molecular selection in potential candidates for anti-EGFR retreatment. In this research framework, we are now conducting the phase II PARERE trial (NCT04787341) randomizing patients previously exposed to first-line anti-EGFR-containing regimens and not harboring RAS/BRAF mutations in their ctDNA to receive panitumumab retreatment followed by regorafenib versus the reverse strategy. The NGS Oncomine™ (10) assay, including 14 genes implied in mCRC progression and anti-EGFR resistance, is used as a screening tool. Here, we present preliminary molecular findings of the screening phase.
2. Materials and methods
2.1. Patient population
RAS/BRAF wild-type mCRC patients enrolled in the molecular screening phase between December 2020 and January 2023 of the randomized phase II PARERE study (NCT04787341) were included. Candidates had to meet the following criteria: diagnosis of unresectable mCRC previously treated with fluoropyrimidines, oxaliplatin, irinotecan, and an antiangiogenic agent (bevacizumab or aflibercept); KRAS/NRAS (codons 12, 13, 59, 61, 117, and 146) and BRAFV600E wt status on primary tumor and/or metastasis; a partial response or stable disease ≥6 months during a previous anti-EGFR-based first-line treatment; ≥4 months interval between the last anti-EGFR administration and LB.
2.2. Sequencing methodology
NGS analysis with Oncomine™ Colon cfDNA Assay was performed at Fondazione IRCSS - Istituto Nazionale dei Tumori di Milano, Milan, Italy. This assay detects frequently mutated single-nucleotide variants (SNVs) and short indels in colon/gastro-intestinal cancers, covering 14 genes with >240 hotspots [Protein Kinase B (AKT1), B-raf Murine Sarcoma Viral Oncogene Homologue B (BRAF), Catenin Beta-1 (CTNNB1), EGFR, Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2), F-box/WD Repeat Containing Protein 7 (FBXW7), Guanine Nucleotide Binding Protein (GNAS), Kirsten Rat Sarcoma Virus (KRAS), Mitogen-activated Protein Kinase Kinase (MAP2K1), Neuroblastoma Ras Viral Oncogene Homologue (NRAS), Phosphatidylinositol 4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha Isoform (PIK3CA), Mothers Against Decapentaplegic (SMAD4), Tumor Protein P53 (TP53), and Adenomatous Polyposis Coli (APC)] with a flexible limit of detection (LOD) of 0.1%–5% that varies with the cfDNA input (1–20 ng). Briefly, sequencing was performed via the use of a tag on plasma samples of circulating free DNA: after attaching a unique molecular tag to the gene-specific primers, the amplified products were grouped into families harboring the same tags. Families containing the same mutant variant were called with optimized Variant Caller settings for the Oncology-Liquid Biopsy application. Families that contained random errors were identified and removed from variant calling. The same test was performed in all samples from patients undergoing molecular screening for the PARERE study.
2.3. Statistical analysis
Patients’ characteristics were described as counts and percentages in the case of categorical variables and as ranges or 95% confidence intervals in the case of continuous variables. Comparisons of the clinical characteristics of the RAS/BRAF wt and mutant groups were performed with the Mann–Whitney, chi-squared, and Fisher’s exact tests, where appropriate. ORR was defined as the ratio between the number of patients achieving at least a partial response, according to Response Evaluation Criteria in Solid Tumors (RECIST) 1.1 criteria (11), and the number of patients who underwent objective radiological assessment at first anti-EGFR exposure. Progression-free survival (PFS) was defined as the interval from the first anti-EGFR exposure to disease progression. LODs of the Oncomine™ panel for KRAS, NRAS, and BRAF mutant clones were compared with the Kruskal–Wallis test and Dunn’s multiple comparison test. Data analysis and visualization were performed with RStudio version 2022.07.02 and Prisma version 4.7.
3. Results
Between December 2020 and January 2023, 218 patients met the eligibility criteria for screening, and ctDNA genotyping was successful in 201 (92%) cases, with a median LOD of 0.11%. KRAS, NRAS, or BRAFV600E mutations were found in 68 (34%) patients. Co-mutations were reported in 14 cases, including 8 cases of KRAS/NRAS mutations (11%), 5 cases of KRAS/BRAFV600E mutations (7%), and 1 case of KRAS/NRAS/BRAFV600E mutations (1%). A total of 133 patients were RAS and BRAFV600E wt and were thus eligible for randomization in the PARERE trial ( Figure 1 ).
Figure 1.
ctDNA screening results according to successful sequencing, RAS/BRAF genotyping and other mutations in the Oncomine panel.
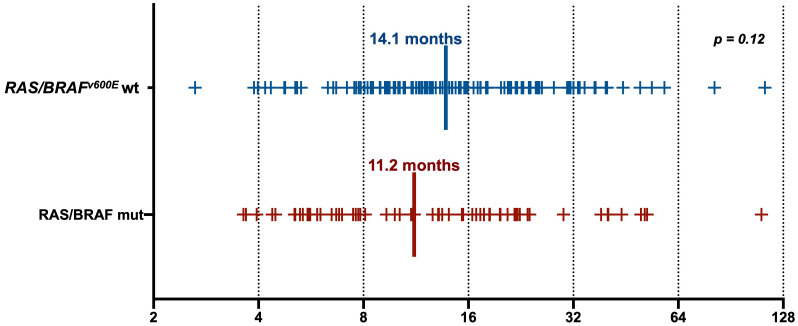
Clinical characteristics of patients with RAS/BRAFV600E wt versus RAS/BRAFV600E mutated ctDNA are summarized in Table 1 . None of them was predictive of ctDNA status at screening for anti-EGFR retreatment, except for a higher percentage of patients with liver-limited disease at the time of the previous anti-EGFR exposure in the RAS/BRAF wt ctDNA group (41% versus 24%, p = 0.02). Notably, the interval between the last anti-EGFR administration and the time of screening was not statistically different between the RAS/BRAFV600E wt and the RAS/BRAFV600E mutated cohorts (14.1 versus 11.2 months, p = 0.12) ( Figure 2 ).
Table 1.
Patients’ characteristics according to ctDNA molecular status by PARERE screening.
| Characteristics | RAS/BRAFV600E wt | RAS/BRAFV600E mut | p | |
|---|---|---|---|---|
| n = 133 (%) | n = 68 (%) | |||
| Sex | Male | 74 (56) | 38 (56) | 0.97 |
| Female | 59 (44) | 30 (44) | ||
| Primary tumor location | Left colon and rectum | 120 (90) | 63 (93) | 0.57 |
| Right colon | 13 (10) | 5 (7) | ||
| Surgery on primary tumor | Yes | 107 (81) | 51 (75) | 0.37 |
| No | 26 (19) | 17 (25) | ||
| Mucinous histology | Yes | 8 (6) | 4 (6) | 1.00 |
| No | 104 (78) | 61 (90) | ||
| NA | 21 (16) | 3 (4) | ||
| MSI status | pMMR or MSS/MSI-low | 131 (99) | 68 (100) | 0.55 |
| dMMR or MSI-high | 2 (1) | 0 (0) | ||
| Time to metastases | >3 months (metachronous) | 31 (23) | 19 (28) | 0.49 |
| ≤3 months (synchronous) | 101 (76) | 49 (72) | ||
| NA | 1 (1) | 0 (0) | ||
| Liver-limited disease at the start of previous anti-EGFR-based treatment | Yes | 54 (41) | 16 (24) | 0.02 |
| No | 79 (59) | 52 (76) | ||
| Peritoneal metastases at the time of LB screening | Yes | 27 (20) | 13 (19) | 0.84 |
| No | 106 (80) | 55 (81) | ||
| Number of metastatic sites at the time of LB screening | 1 | 19 (14) | 8 (12) | 0.21 |
| 2 | 48 (36) | 23 (34) | ||
| 3 | 50 (38) | 21 (31) | ||
| > 3 | 16 (12) | 16 (23) | ||
| Previous anti-EGFR-based treatment regimen | FOLFIRI + anti-EGFR | 29 (22) | 16 (23) | 0.73 |
| FOLFOX/XELOX + anti-EGFR | 78 (59) | 43 (63) | ||
| FOLFOXIRI + anti-EGFR | 19 (14) | 7 (10) | ||
| Anti-EGFR ± monochemotherapy | 7 (5) | 2 (3) | ||
| Previous anti-EGFR-based treatment response | CR/PR | 111 (84) | 60 (88) | 0.37 |
| SD | 22 (16) | 8 (12) | ||
| mPFS to previous anti-EGFR exposure | Median, months | 13.1 | 14.3 | 0.95 |
| (95% CI) | (12.3–63.6) | (12.0–67.1) | ||
| Time interval between last anti-EGFR administration and disease progression | >3 months | 62 (47) | 39 (57) | 0.15 |
| ≤3 months | 71 (53) | 29 (43) | ||
| Time interval between last anti-EGFR exposure and LB screening | Median, months | 14.1 | 11.2 | 0.12 |
| (95% CI) | (12.3–15.7) | (9.9–15.5) | ||
| Lines of treatment between last anti-EGFR exposure and LB screening | None | 1 (1) | 1 (1) | 0.75 |
| 1 | 98 (74) | 50 (74) | ||
| 2 | 26 (20) | 14 (21) | ||
| 3 | 6 (4) | 1 (1) | ||
| 4 | 2 (1) | 2 (3) |
MSI, microsatellite instability; anti-EGFR, anti-epidermal growth factor receptor monoclonal antibodies; CI, confidence interval; CR, complete response; LB, liquid biopsy; NA, not available; PFS, progression-free survival; PR, partial response, SD, stable disease; ctDNA, circulating tumor DNA; pMMR, proficient mismatch repair; dMMR, deficient mismatch repair.
Bolded value corresponds to a statically significant p-value as compared with the others (not statistically significant).
Figure 2.
Anti-EGFR free interval in ctDNA RAS/BRAFV600E wild-type and mut patients.
The LOD was superimposable in patients with RAS/BRAFV600E wt and RAS/BRAFV600E mutant clones (0.12% versus 0.10%, respectively, p = 0.46) ( Supplementary Figure 1 ). The distribution of variant allele fraction (VAF) among KRAS, NRAS, and BRAF mutant tumors was not homogeneous (p = 0.01), with BRAF mutant clones showing a lower VAF compared to KRAS (0.19% versus 0.52%, p = 0.009) and NRAS mutant clones (0.19 versus 0.62%, p = 0.07) ( Supplementary Figure 2 ). Among KRAS alterations, the most frequent driver of secondary resistance to anti-EGFRs was KRASQ61H mutation (31%), followed by KRASG12D (21%) and KRASG12A (16%). The druggable KRASG12C mutation was found in 3% of patients with KRAS mutant ctDNA ( Supplementary Figure 3 ).
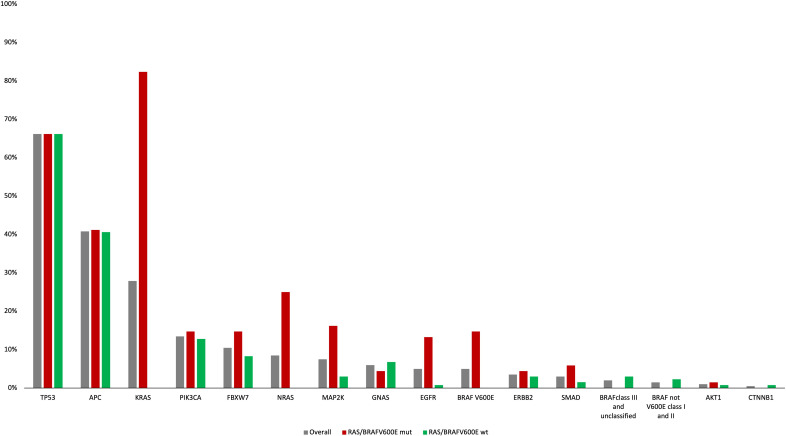
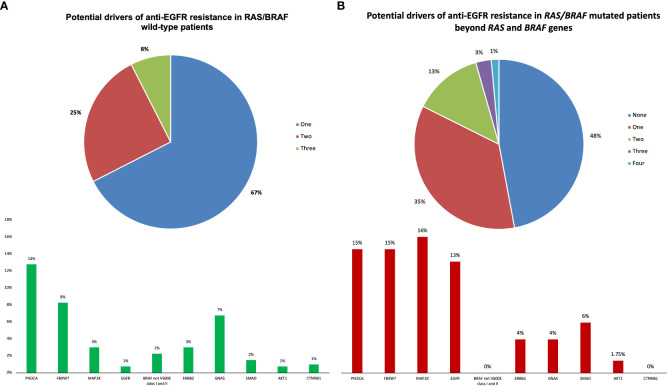
Among RAS/BRAFV600E wt patients, 28 (21%) did not harbor any mutation included in the Oncomine™ panel. Among the others, mutations in founder genes such as TP53 and APC were detected in 88 (66%) and 53 (40%) patients, respectively, with frequent co-mutations. Most importantly, 40 out of 133 RAS/BRAFV600E wt patients (30%) had a mutation in at least another gene potentially implied in anti-EGFR resistance—including class I and II non-BRAFV600E mutations ( Figure 3 ). Notably, 27 patients (67%) harbored one single potential driver of resistance, with co-mutations in two or three genes occurring in 10 (25%) and 3 (8%) cases, respectively. In detail, mutations in PIK3CA, FBXW7, GNAS, MAP2K1, ERBB2, BRAF (class I and II non-BRAFV600E), SMAD, EGFR, AKT1, and CTNNB1 occurred in 13%, 8%, 7%, 3%, 2%, 2%, 1%, 1%, 1%, and 1% of patients, respectively. Of 68 patients, 36 (53%) with RAS and/or BRAFV600E mutant clones showed the co-presence of other drivers of resistance as well ( Figure 4A ). Overall, one, two, three, and four other potential drivers of resistance to anti-EGFRs were found in 67%, 25%, 6%, and 1% of patients, respectively. The relative frequency of these co-mutations was different compared to that of RAS/BRAFV600E wt patients, with a higher frequency of MAP2K1 (16%), FBXW7 (15%), EGFR (13%), and SMAD (6%) mutations ( Figure 4B ).
Figure 3.
Frequency of genomic alterations according to the Oncomine panel in the overall, RAS/BRAFV600E mut and RAS/BRAFV600E wt populations.
Figure 4.
Potential drivers of anti-EGFR resistance beyond RAS/BRAF genes in RAS/BRAF wild-type and mutated patients.
The median VAF of altered genes other than RAS and BRAFV600E driving potential resistance to anti-EGFRs was subclonal and numerically higher in the RAS/BRAFV600E wt group compared to the RAS/BRAFV600E mutant (0.56% versus 0.23%, p = 0.22) ( Supplementary Figure 4 ).
4. Discussion
A growing number of therapeutic options have recently become available in the chemorefractory landscape of mCRC patients, and others are under investigation, including anti-EGFR retreatment in patients with RAS/BRAF wt tumors (1, 10, 12–14). In this regard, the most convincing evidence has been collected in patients selected according to the analysis of ctDNA by means of high-throughput sequencing technologies (i.e., digital droplet PCR) aimed at revealing the lowest fraction of resistant clones in the blood while limiting the analysis to a restricted number of genes (i.e., RAS, BRAF, and EGFR) (1, 4, 5). In patients with RAS, BRAF, and EGFR wt ctDNA, according to digital droplet PCR (ddPCR), the anti-EGFR retreatment provided a 30% ORR and 4-month PFS (1, 4, 5). The positioning of anti-EGFR retreatment in the current landscape of therapeutic options for chemorefractory mCRC patients will be likely clarified by evidence from randomized trials. This is the objective of the PARERE study: to adopt an NGS technique as a screening tool and to clarify in a post-hoc analysis the impact of other alterations of acquired resistance.
Our series is the largest prospective cohort (n = 202) of patients candidates for anti-EGFR retreatment undergoing molecular screening in LB. The RAS/BRAFV600E conversion rate of 34% reported here mirrors previous data from the CHRONOS trial (29%) obtained with a digital droplet PCR assay (1), suggesting that the flexible LOD of the NGS approach used in the PARERE study still provides the advantage of a massively parallel sequencing while preserving an adequate sensitivity to detect RAS/BRAF mutant clones (1). KRASQ61H and KRASG12A hotspot mutations, respectively accounting for 35% and 16% of identified KRAS mutations, were much more frequent than in the chemo-naïve setting where their incidence is approximately 1.5% and 4%, respectively, among KRAS mutant tumors (15–17). Our data corroborate previous findings by Woolston et al. and highlight that KRAS mutations selected under the pressure of EGFR blockade follow different mechanisms of molecular adaptation than those selected in earlier stages of the disease (18). From a biochemical standpoint, KRAS codon 61 mutations yield higher in vitro KRAS-GTP levels as compared to codon 12 and 13 mutations and share with KRAS G12A a lower intrinsic GTPase activity and a stronger affinity for RAF gene, as compared to mutations in other codons (19, 20). These considerations may be relevant in the current and future research efforts toward mutation-specific and pan-KRAS inhibitors (21, 22).
Notably, no clinical characteristics were reliable in predicting RAS/BRAFV600E status (1). Numerically longer anti-EGFR-free intervals and time from the last anti-EGFR administration to first-line disease progression were observed in the group with RAS/BRAFV600E wt ctDNA. Translational data from the randomized FIRE4 trial showed a significant correlation between the duration of the exposure to first-line anti-EGFRs and the occurrence of RAS mutations, though in the absence of any overall survival (OS) difference between the groups of patients acquiring or not RAS mutations in ctDNA (23).
We managed to detect rare subclonal alterations potentially implied in anti-EGFR resistance in the 30% of RAS/BRAFV600E wt patients, with co-mutations occurring in roughly one-third of them, providing further evidence that co-evolution of different subclones is a common mechanism of escape to targeted therapy, as recently observed in patients with RAS wt tumors treated with an anti-EGFR and then enrolled in the CO.26 trial (24) and in BRAFV600E mutant mCRC patients treated with encorafenib plus cetuximab in the BEACON trial (25). Among RAS and BRAF wt ctDNA patients, potential drivers of anti-EGFR resistance were found in 28% of subjects, mostly including PIK3CA, FBXW7, and GNAS mutations (28%), whereas AKT, EGFR, SMAD, and CTNNB1 mutations were rare (4%). Remarkably, patients with RAS/BRAFV600E mutations in their ctDNA showed a different genomic pattern, with redundant activation of the MAPK pathway through EGFR and MAP2K1 mutations in 29% of patients.
Our work suffers from several limitations. First, blood samples were collected and analyzed only at the time of screening for the PARERE study and not at the diagnosis of mCRC, thus not allowing for full exclusion of RAS/BRAF mutations that could be detected in ctDNA before the first-line anti-EGFR-based therapy, though in the absence of mutations in tissue DNA, as described in up to 10% of RAS/BRAFV600E wt mCRC patients (26). Second, the Oncomine™ panel does not allow the identification of molecular alterations other than SNVs (i.e., amplifications or rearrangements) and covers only 14 genes; other drivers of anti-EGFR resistance may have been missed at screening. Third, only outcome data from the PARERE trial will clarify the impact of these alterations on the clinical activity of the anti-EGFR retreatment.
In conclusion, preliminary molecular findings from the PARERE study corroborate the need to use LB as a screening tool before offering anti-EGFR retreatment, considering the lack of reliability of potential clinical surrogates of EGFR dependency. The subclonality of molecular alterations identified in these patients makes quite challenging the idea of exploiting these events as efficacious targets for subsequent tailored treatments.
Data availability statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA1064885.
Ethics statement
The studies involving humans were approved by Comitato Etico Regione Toscana - Area Vasta Nord Ovest (CEAVNO) - Codice OsSC: OCE000000056 - ORG ID: ORG-100032439. The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MMG: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing, Supervision, Visualization, Validation. GV: Data curation, Formal Analysis, Investigation, Writing – original draft, Validation. MG: Data curation, Writing – review & editing, Investigation. PC: Data curation, Formal Analysis, Investigation, Writing – original draft, Writing – review & editing, Validation. IC: Data curation, Writing – review & editing, Investigation. ET: Data curation, Writing – review & editing, Investigation. EC: Data curation, Writing – review & editing, Investigation. ABu: Data curation, Writing – review & editing, Investigation. FP: Data curation, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision, Validation. VP: Data curation, Writing – review & editing, Validation. ABo: Data curation, Writing – review & editing, Validation. FS: Data curation, Writing – review & editing, Validation. MB: Data curation, Writing – review & editing, Validation. PM: Data curation, Writing – review & editing, Validation. SL: Data curation, Writing – review & editing, Funding acquisition, Project administration, Resources, Supervision, Validation. VC: Data curation, Writing – review & editing, Validation. BB: Data curation, Writing – review & editing, Validation. MC: Data curation, Writing – review & editing, Validation. MR: Writing – review & editing, Investigation, Supervision, Validation. GF: Writing – review & editing, Investigation, Supervision, Validation. DR: Data curation, Formal Analysis, Writing – review & editing, Conceptualization, Investigation, Supervision, Validation, Writing – original draft. CC: Conceptualization, Data curation, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing, Formal Analysis, Funding acquisition, Project administration, Resources, Validation.
Acknowledgments
The research leading to these results has received funding from the European Union - NextGenerationEU through the Italian Ministry of University and Research under PNRR - M4C2-I1.3 Project PE_00000019 “HEAL ITALIA” to CC and GF CUP: I53C22001440006. The views and opinions expressed are those of the authors only and do not necessarily reflect those of the European Union or the European Commission. Neither the European Union nor the European Commission can be held responsible for them.
Funding Statement
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The study was supported by GONO and ARCO Foundations, Amgen, and Bayer (no grant number applicable). The sponsors had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Conflict of interest
SL: consulting or an advisory role—Amgen, Merck Serono, Lilly, AstraZeneca, Incyte, Daiichi-Sankyo, BMS, Servier, and MSD; research funding—Amgen, Merck Serono, Bayer, Roche, Lilly, AstraZeneca, and BMS; speakers’ fees from Roche, Lilly, BMS, Servier, Merck Serono, Pierre-Fabre, GlaxoSmithKline, and Amgen. FP: honoraria—Merck-Serono, Amgen, Bayer, Servier, Takeda, Pierre-Fabre, MSD, BMS, and Astellas; research grants for non-profit studies—AstraZeneca, BMS, Agenus, Amgen. CC: honoraria—Amgen, Bayer, Merck, Roche, and Servier; consulting or advisory role—Amgen, Bayer, MSD, and Roche; speakers’ bureau—Servier; research funding—Bayer, Merck, and Servier; travel, accommodations, and expenses—Roche and Servier. DR: honoraria—Takeda Pharmaceuticals.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fonc.2023.1307545/full#supplementary-material
References
- 1. Sartore-Bianchi A, Pietrantonio F, Lonardi S, Mussolin B, Rua F, Crisafulli G, et al. Circulating tumor DNA to guide rechallenge with panitumumab in metastatic colorectal cancer: the phase 2 CHRONOS trial. Nat Med (2022) 28(8):1612–8. doi: 10.1038/s41591-022-01886-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Nakamura M. Phase II study of cetuximab rechallenge in patients with RAS wild-type metastatic colorectal cancer: E-rechallenge trial. Ann Oncol (2019) 30:vi116. doi: 10.1093/annonc/mdz338.112 [DOI] [Google Scholar]
- 3. Sunakawa Y, Nakamura M, Ishizaki M, Kataoka M, Satake H, Kitazono M, et al. RAS mutations in circulating tumor DNA and clinical outcomes of rechallenge treatment with anti-EGFR antibodies in patients with metastatic colorectal cancer. JCO Precis. Oncol (2020) 4):898–911. doi: 10.1200/PO.20.00109 [DOI] [PubMed] [Google Scholar]
- 4. Parseghian CM, Vilar Sanchez E, Sun R, Eluri M, Morris VK, Johnson B, et al. Phase 2 study of anti-EGFR rechallenge therapy with panitumumab with or without trametinib in advanced colorectal cancer. J Clin Oncol (2022) 40(16_suppl):3520–0. doi: 10.1200/JCO.2022.40.16_suppl.3520 [DOI] [Google Scholar]
- 5. Kagawa Y, Kotani D, Bando H, Takahashi N, Hamaguchi T, Kanazawa A, et al. Plasma RAS dynamics and anti-EGFR rechallenge efficacy in patients with RAS/BRAF wild-type metastatic colorectal cancer: REMARRY and PURSUIT trials. J Clin Oncol (2022) 40(16_suppl):3518–8. doi: 10.1200/JCO.2022.40.16_suppl.3518 [DOI] [Google Scholar]
- 6. Cremolini C, Rossini D, Dell’Aquila E, Lonardi S, Conca E, Del Re M, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: A phase 2 single-arm clinical trial. JAMA Oncol (2019) 5(3):343–50. doi: 10.1001/jamaoncol.2018.5080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rossini D, Germani MM, Pagani F, Pellino A, Dell'Aquila E, Bensi M, et al. Retreatment with anti-EGFR antibodies in metastatic colorectal cancer patients: A multi-institutional analysis. Clin Colorectal Cancer (2020) 19(3):191–199.e6. doi: 10.1016/j.clcc.2020.03.009 [DOI] [PubMed] [Google Scholar]
- 8. Schulz MS, Wolf S, Struck V, Thomas N, Husman G, Zeuzem S, et al. Anti-EGFR reintroduction and rechallenge in metastatic colorectal cancer (mCRC): A real-world analysis. Cancers (2022) 14(7):1641. doi: 10.3390/cancers14071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mariani S, Puzzoni M, Giampieri R, Ziranu P, Pusceddu V, Donisi C, et al. Liquid biopsy-driven cetuximab rechallenge strategy in molecularly selected metastatic colorectal cancer patients. Front Oncol (2022) 12:852583. doi: 10.3389/fonc.2022.852583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moretto R, Rossini D, Capone I, Boccaccino A, Perrone F, Tamborini E, et al. Rationale and study design of the PARERE trial: randomized phase II study of panitumumab re-treatment followed by regorafenib versus the reverse sequence in RAS and BRAF wild-type chemo-refractory metastatic colorectal cancer patients. Clin Colorectal Cancer (2021) 20(4):314–7. doi: 10.1016/j.clcc.2021.07.001 [DOI] [PubMed] [Google Scholar]
- 11. Schwartz LH, Litière S, de Vries E, Ford R, Gwyther S, Mandrekar S, et al. RECIST 1.1 – update and clarification: from the RECIST committee. Eur J Cancer Oxf. Engl 1990 (2016) 62:132–7. doi: 10.1016/j.ejca.2016.03.081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dasari A, Lonardi S, Garcia-Carbonero R, Elez E, Yoshino T, Sobrero A, et al. Fruquintinib versus placebo in patients with refractory metastatic colorectal cancer (FRESCO-2): an international, multicentre, randomised, double-blind, phase 3 study. Lancet (2023) 402(10395):41–53. doi: 10.1016/S0140-6736(23)00772-9 [DOI] [PubMed] [Google Scholar]
- 13. Prager GW, Taieb J, Fakih M, Ciardiello F, Van Cutsem E, Elez E, et al. Trifluridine–tipiracil and bevacizumab in refractory metastatic colorectal cancer. N Engl J Med (2023) 388(18):1657–67. doi: 10.1056/NEJMoa2214963 [DOI] [PubMed] [Google Scholar]
- 14. Bullock A, Grossman J, Fakih M, Lenz H, Gordon M, Margolin K, et al. LBA O-9 Botensilimab, a novel innate/adaptive immune activator, plus balstilimab (anti-PD-1) for metastatic heavily pretreated microsatellite stable colorectal cancer. Ann Oncol (2022) 33:S376. doi: 10.1016/j.annonc.2022.04.453 [DOI] [Google Scholar]
- 15. Neumann J, Zeindl-Eberhart E, Kirchner T, Jung A. Frequency and type of KRAS mutations in routine diagnostic analysis of metastatic colorectal cancer. Pathol - Res Pract (2009) 205(12):858–62. doi: 10.1016/j.prp.2009.07.010 [DOI] [PubMed] [Google Scholar]
- 16. Imamura Y, Lochhead P, Yamauchi M, Kuchiba A, Qian ZR, Liao X, et al. Analyses of clinicopathological, molecular, and prognostic associations of KRAS codon 61 and codon 146 mutations in colorectal cancer: cohort study and literature review. Mol Cancer (2014) 13(1):135. doi: 10.1186/1476-4598-13-135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Prior IA, Hood FE, Hartley JL. The frequency of ras mutations in cancer. Cancer Res (2020) 80(14):2969–74. doi: 10.1158/0008-5472.CAN-19-3682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Woolston A, Barber LJ, Griffiths B, Pich O, Lopez-Bigas N, Matthews N, et al. Mutational signatures impact the evolution of anti-EGFR antibody resistance in colorectal cancer. Nat Ecol Evol (2021) 5(7):1024–32. doi: 10.1038/s41559-021-01470-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stolze B, Reinhart S, Bulllinger L, Fröhling S, Scholl C. Comparative analysis of KRAS codon 12, 13, 18, 61, and 117 mutations using human MCF10A isogenic cell lines. Sci Rep (2015) 5:8535. doi: 10.1038/srep08535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hunter JC, Manandhar A, Carrasco MA, Gurbani D, Gondi S, Westover KD. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res (2015) 13(9):1325–35. doi: 10.1158/1541-7786.MCR-15-0203 [DOI] [PubMed] [Google Scholar]
- 21. Kim D, Herdeis L, Rudolph D, Zhao Y, Böttcher J, Vides A, et al. Pan-KRAS inhibitor disables oncogenic signalling and tumour growth. Nature (2023) 619(7968):160–6. doi: 10.1038/s41586-023-06123-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Punekar SR, Velcheti V, Neel BG, Wong K-K. The current state of the art and future trends in RAS-targeted cancer therapies. Nat Rev Clin Oncol (2022) 19(10):637–55. doi: 10.1038/s41571-022-00671-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Stintzing S, Fischer von Weikersthal L, Fuchs M, Fuchs M, Kaiser F, Heinrich K, Modest DP, et al. Phase III FIRE-4 study (AIO KRK-0114): Evaluation of first-line treatment efficacy of FOLFIRI/cetuximab in patients with RAS-WT mCRC receiving the first cycle of treatment with chemotherapy only. J Clin Oncol (2023) 41(4_suppl):100–0. doi: 10.1200/JCO.2023.41.4_suppl.100 [DOI] [Google Scholar]
- 24. Topham JT, O’Callaghan CJ, Feilotter H, Kennecke HF, Lee YS, Li W, et al. Circulating tumor DNA identifies diverse landscape of acquired resistance to anti–epidermal growth factor receptor therapy in metastatic colorectal cancer. J Clin Oncol (2023) 41(3):485–96. doi: 10.1200/JCO.22.00364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kopetz S, Murphy DA, Pu J, Yaeger R, Ciardiello F, Desai J, et al. 316O Genomic mechanisms of acquired resistance of patients (pts) with BRAF V600E-mutant (mt) metastatic colorectal cancer (mCRC) treated in the BEACON study. Ann Oncol (2022) 33:S681–2. doi: 10.1016/j.annonc.2022.07.454 [DOI] [Google Scholar]
- 26. Manca P, Corallo S, Busico A, Lonardi S, Corti F, Antoniotti C, et al. The added value of baseline circulating tumor DNA profiling in patients with molecularly hyperselected, left-sided metastatic colorectal cancer. Clin Cancer Res Off J Am Assoc Cancer Res (2021) 27(9):2505–14. doi: 10.1158/1078-0432.CCR-20-4699 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ncbi.nlm.nih.gov/sra/PRJNA1064885.