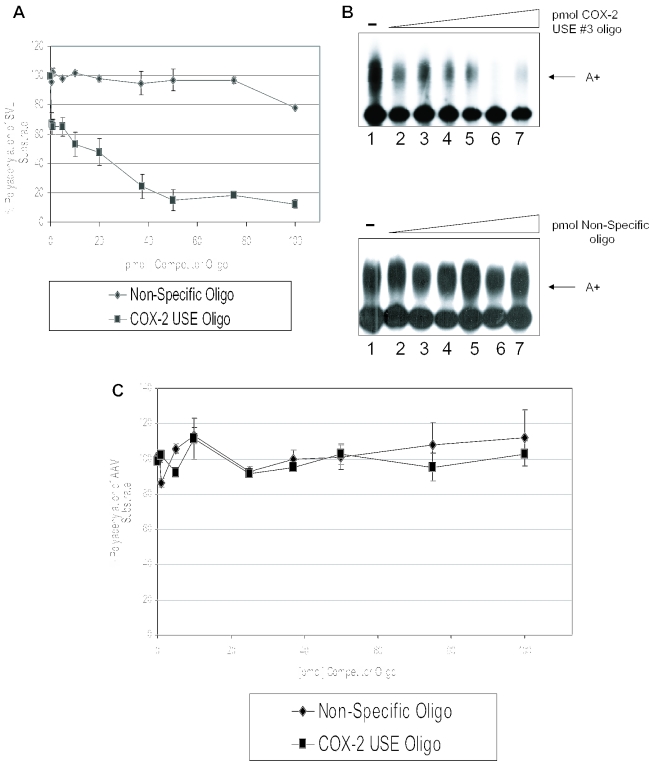
Figure 5.
Competition studies suggest USE binding factors influence the processing efficiency of COX-2 proximal polyadenylation signal. (A) Graphical representation of in vitro competitions using SVL and either COX-2 USE oligoribonucleotide (squares) or non-specific oligoribonucleotide (diamonds). Numbers on the x-axis represent pmol of competitor added, numbers on the y-axis represent percent polyadenylation. This quantification represents the average of three independent experiments. Error bars represent SD. (B) Representative in vitro polyadenylation reactions using SV40 late polyadenylation signal RNA as the substrate RNA and increasing amounts of either COX-2 USE oligoribonucleotide as competitor (upper panel) and non-specific oligoribonucleotide as competitor (lower panel). Amounts of competitor used in both sets of reactions are as follows: lane 1s, no competitor; lane 2s, 1 pmol competitor; lane 3s, 10 pmol; lane 4s, 20 pmol; lane 5s, 37 pmol; lane 6s, 50 pmol; and lane 7s, 75 pmol. (C) Graphical representation of in vitro competitions using AAV substrate RNA and either COX-2 USE oligoribonucleotide (squares) or non-specific oligoribonucleotide (diamonds).