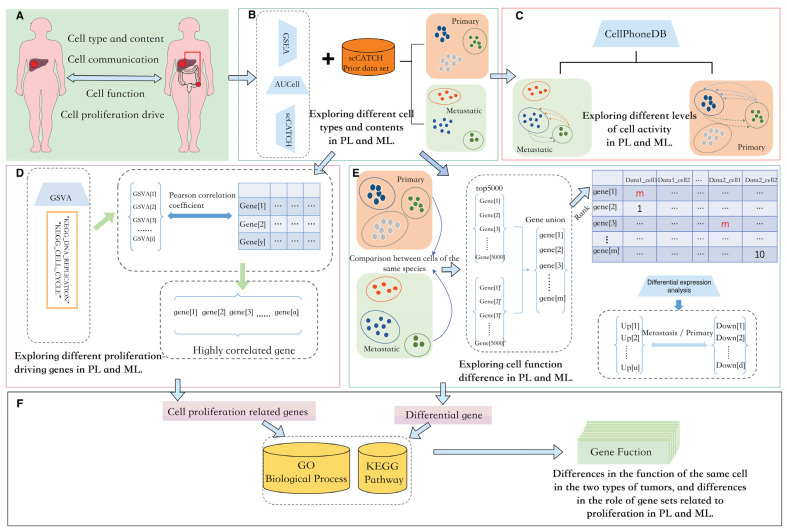
Figure 1.
Overview of our study. (A): We mainly carry out four studies on metastatic liver cancer and primary liver cancer: cell type and content, cell communication, cell function, and cell proliferation. (B): Cell annotation is completed using GSEA, AUCell, and scCATCH paired with a priori gene sets included in scCatch. (C): CellPhoneDB is used to calculate the communication strength of various types of cells. (D): GSVA calculates the enrichment of genes in the tissue in proliferation-related pathways and then we select genes with high Pearson correlation coefficients. (E): Differential analysis of similar cells in two tumor types uses a rank-based approach. (F): The proliferation-related genes and cell differential genes selected in Figures (D,E) are enriched in the GO:BP and KEGG databases. PL: primary liver cancer. ML: metastatic liver cancer.