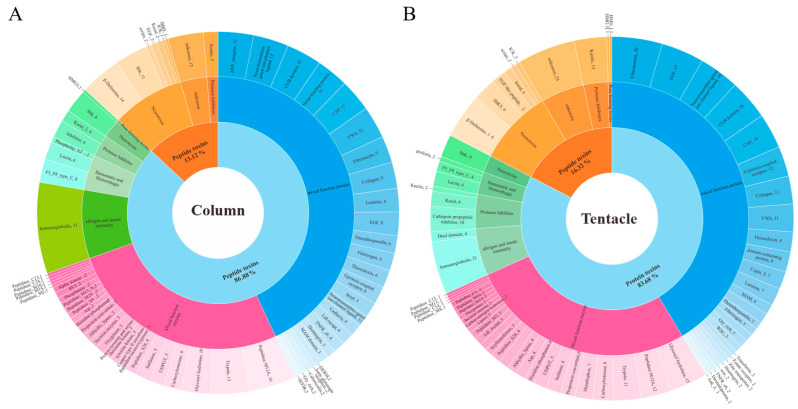
Figure 2.
Putative toxin families identified in the transcriptomes of H. magnifica. The distinctive protein and peptide sequences from the tentacles (442) and the column (381) were compared by blasting the UniProt database (www.UniProt.org/program/toxins, Accessed on: 30 August 2023). They were then classified into toxin families based on their cysteine scaffolds and amino acid sequences. The pie charts illustrate the proportional contribution of each toxin family to the predicted venom proteome. The number of homologs identified for each protein and peptide family is displayed next to the family name. (A) Classification of the tentacles transcriptome; (B) classification of the column transcriptome.