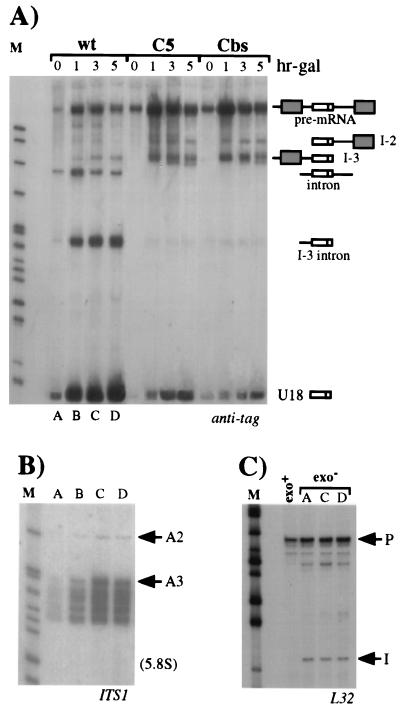
FIG. 5.
Lack of the major 5′→3′ exonucleases does not affect U18 production. (A) Northern analysis of 5 μg of total RNA extracted from the rat1-1 xrn1Δ double mutant strain transformed with the pGALU18wt (wt lanes), pGALU18C5 (C5 lanes), or pGALU18Cbs (Cbs lanes) construct (Fig. 1). Cultures were shifted to the nonpermissive temperature, and after 1.5 h, galactose was added to the medium (hr-gal). Hybridization was performed with the anti-tag oligonucleotide. The different products are indicated on the right as in Fig. 2. The numbers above the lanes indicate hours of galactose induction. (B) Samples (5 μg) of the same RNAs extracted from the rat1-1 xrn1Δ strain transformed with the pGALU18wt construct (lanes A through D of panel A) were electroblotted onto a separate filter and hybridized with an oligonucleotide complementary to the ITS1 region of rRNA (see Materials and Methods). The positions of the two pre-5.8S rRNA species are indicated on the right. The migration of the mature 5.8S rRNA is also indicated. (C) Primer extension analysis using oligonucleotide L32 (see Materials and Methods), which is specific for the intronic sequence of the RPL32 transcript, performed on the same RNA preparations from the rat1-1 xrn1Δ strain (exo− lanes) transformed with the pGALU18wt construct (lanes A, C, and D of panel A) and on control RNA extracted from strain CH1462 (exo+ lane). The extended products corresponding to the 5′ ends of the intron (I) and of the pre-mRNA (P) are indicated. Although not shown, DNA sequencing reactions using oligonucleotide L32 were run alongside the primer extension reactions. Lanes M contained MspI-digested pBR322 plasmid DNA.