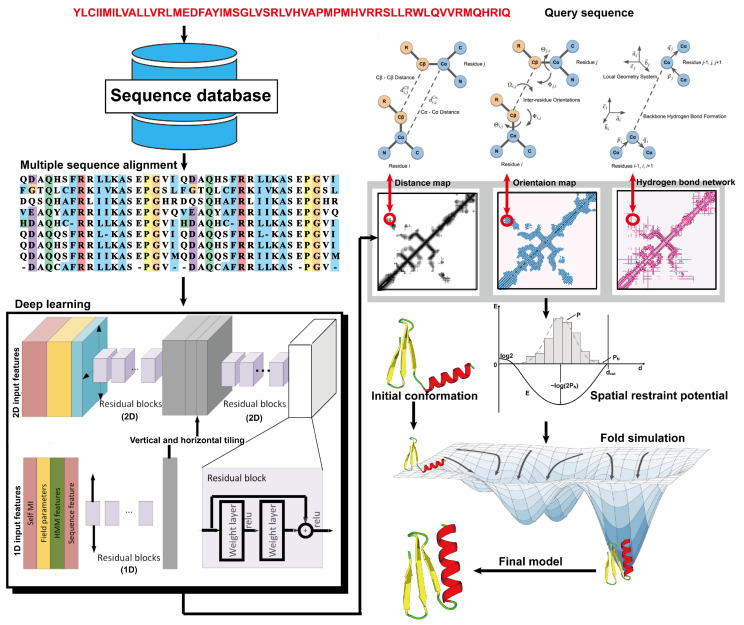
Figure 5.
Illustration of distance-based protein structure prediction methods. Starting from a query sequence, an MSA is first generated by searching through databases. Then, the MSA is fed into deep neural networks to predict spatial restraints, such as distance maps, inter-residue orientations, and hydrogen bond networks. Finally, the final structural model is constructed by employing the potentials extracted from the predicted spatial restraints in a folding simulation to identify the lowest energy structure.