Figure 5.
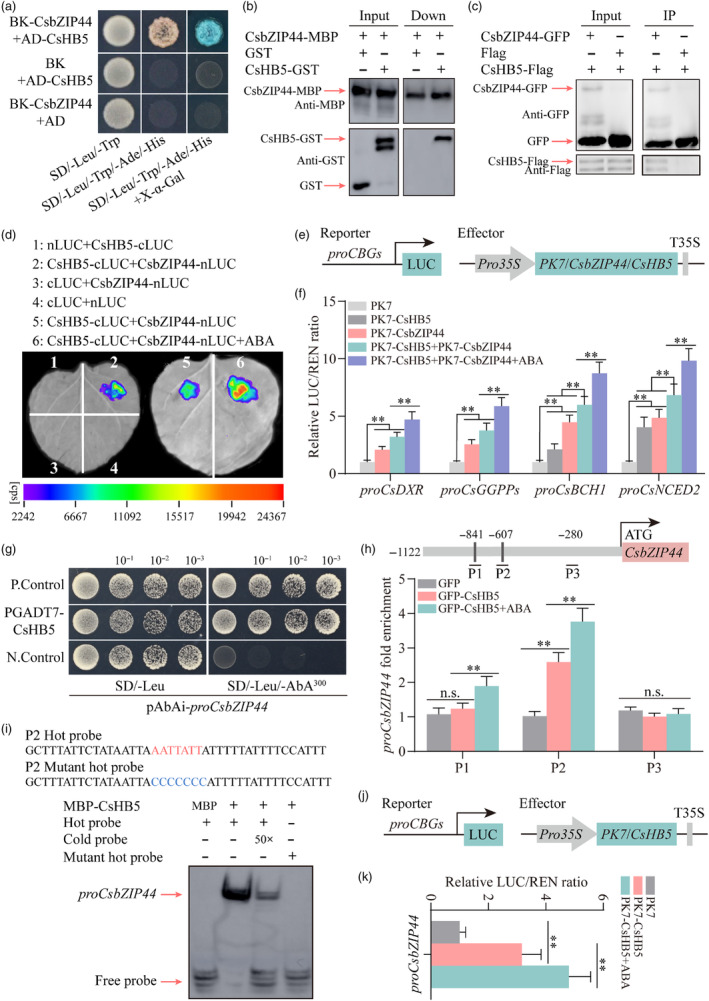
CsHB5 interacts with CsbZIP44 and transcriptionally activates CsbZIP44. (a) Yeast two‐hybrid (Y2H) assay revealing an interaction between CsbZIP44 and CsHB5. Yeast grown in SD/−Trp/−Leu medium and SD/−Trp/−Leu/−His/−Ade medium is shown. The interaction is indicated by yeast growth and X‐α‐Gal staining. (b) The interactions between CsbZIP44 and CsHB5 were analysed using a pull‐down assay. Fusion proteins GST‐CsHB5 and MBP‐CsbZIP44 were used in the pull‐down analysis. GST‐ and MBP‐antibodies were used for immunoblot analyses. The band detected by the GST antibody in the pull‐down protein sample indicates the interaction between CsbZIP44 and CsHB5. (c) The interaction between CsbZIP44 and CsHB5 was confirmed with a co‐immunoprecipitation (Co‐IP) assay. The fused constructs GFP‐tagged CsbZIP44, and flag‐tagged CsHB5 were co‐overexpressed in Nicotiana benthamiana leaves. GFP antibody beads were used for immunoprecipitation. GFP‐ and flag‐antibodies were used for immunoblot analyses. The band detected by the GFP antibody in the IP samples indicates an interaction between CsbZIP44 and CsHB5. (d) A luciferase complementation imaging assay shows that CsbZIP44 interacts with CsHB5, and this interaction is enhanced by ABA treatment. Agrobacterium tumefaciens strain GV3101 harbouring different constructs was infiltrated into different wild tobacco leaf regions. Luciferase activities were imaged in these regions 3 days after infiltration. cps, signal counts per second. (e) Schematic representation of reporter and effector constructs used in dual‐luciferase assay. (f) Dual‐luciferase assay showing that the interaction between CsbZIP44 and CsHB5 significantly increases the activation effect on the promoter activity of target genes, which is strengthened by ABA treatment. (g) Y1H assay showing interactions of CsHB5 with CsbZIP44 promoter. Empty PGADT7 + pAbAi‐proCsbZIP44 and PGADT7‐Rec‐p53 + p53‐AbAi as the negative (N. Control) and positive controls (P. Control) respectively. Aureobasidin A (AbA) is a yeast cell growth inhibitor. SD/−Leu/AbA250, SD/−Leu medium supplemented with 250 ng ml−1 as the basal concentration of proCsbZIP44. (h) ChIP‐PCR assay indicating the interaction of CsHB5 with several regions in the CsbZIP44 promoter in vivo. The grey lines represent putative binding motif of HD‐ZIP family proteins in the CsbZIP44 promoter. Cross‐linked chromatin samples were extracted from GFP‐CsHB5 fruit calli treated with or without ABA (250 μm) and precipitated with an anti‐GFP antibody. Eluted DNA was used to amplify the sequences by q‐PCR. (i) Electrophoretic mobility shift assay (EMSA) assay confirming that CsHB5 directly binds the binding element of HD‐ZIP TFs in the CsbZIP44 promoter in vitro. Purified MBP‐tagged CsHB5 protein was used in EMSA assay, and purified MBP protein was used as a negative control. Black arrows indicate the position of biotin‐labelled promoter fragment (hot probe) containing the putative binding motifs of HD‐ZIP family proteins. Red arrows indicate the positions of protein‐DNA complexes or free probes. Red letters represent the binding motifs, and blue letters indicate their corresponding mutant motifs. ‘+’ and ‘–’ the presence and absence of the indicated probe or protein respectively. Increasing amounts (50 folds) of the unlabelled DNA fragments (cold probe) were added as competitors. (j) Schematic representation of reporter and effector constructs used in dual‐luciferase assay. (k) Dual‐luciferase assay showing that CsHB5 enhances the activation of the CsbZIP44 promoter, and this activation is induced by ABA treatment. ABA treatment (100 μm) in the luciferase complementation imaging assay and the dual‐luciferase assay was conducted for 3 h before determination. Data represent means ± SD of three biological replicates. Asterisks indicate statistically significant differences determined by Student's t‐test (*, 0.01 < P < 0.05; **, P < 0.01; n.s., no significant difference).
