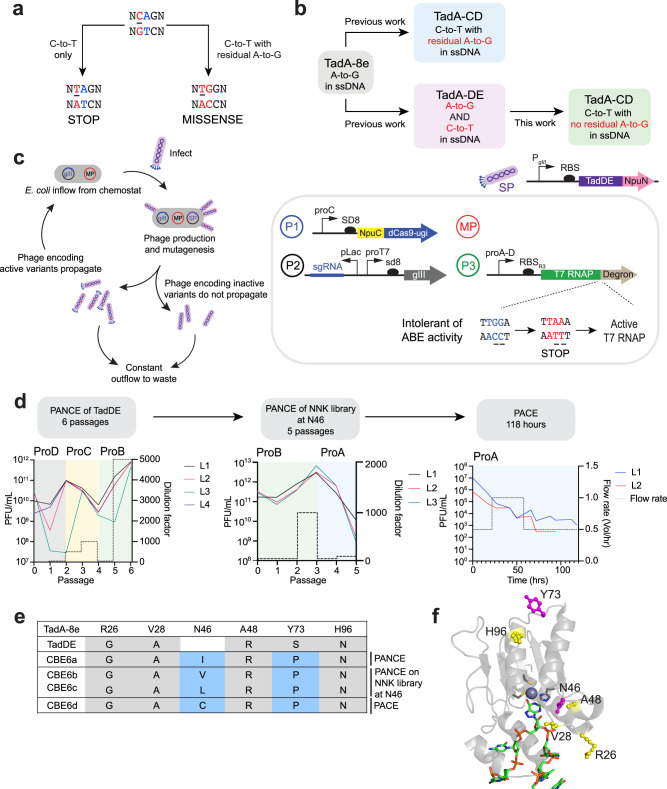
Fig. 1. Development of a highly active and selective cytosine base editor from a TadA dual base editor using phage-assisted evolution.
a An active and selective cytosine base editor, but not one with residual adenine base editing activity, can cleanly install stop codons into target genes. b Schematic of the evolution of a cytosine base editor from a TadA-derived dual base editor (TadDE)29. c Diagram depicting phage-assisted continuous evolution (PACE, left) and the selection circuit used in this study (right). A continuous flow of E. coli host cells with the selection circuit and a mutagenesis plasmid (red) are infected by selection phage encoding a deaminase (SP). In the selection circuit, phage propagation is linked with gIII expression (P2), which can only be transcribed with active T7 RNA polymerase. T7 RNA polymerase (P3) is fused to a C-terminal degron, and the deaminase must perform C•G-to-T•A editing to install a stop codon before the degron to generate active T7 RNA polymerase. In the event of phage infection, the full base editor is reconstituted using a split-intein system (P1), and mutations accumulate in the deaminase. Beneficial mutations lead to phage propagation and enrichment in the lagoon, while the less-fit phage are unable to propagate and are washed out by the constant outflow. d Evolutionary trajectory of an active and selective cytosine base editor from TadDE. Phage-assisted non-continuous evolution (PANCE) was performed on TadA-DE until phage titers increased despite higher stringency. The resulting genotypes identified a conserved mutation at position N46 in TadA, so an NNK library was constructed to diversify this position, and PANCE was performed on the resulting variants. PACE was performed for >100 hrs on the resulting variants from both PANCE experiments. Dilution factors are indicated on the right y-axis. Relative promoter units (normalized to proD) for proA, proB, proC, and proD are as follows: 0.030, 0.119, 0.278, and 1.000, respectively55. e Mutation table from evolved deaminases showing conserved mutations. f Cryo-EM structure of ABE8e (PDB: 6VPC) with mutations labeled. New mutations are highlighted in magenta, and mutations inherited from TadDE are highlighted in yellow. Source data are provided as a Source Data file.