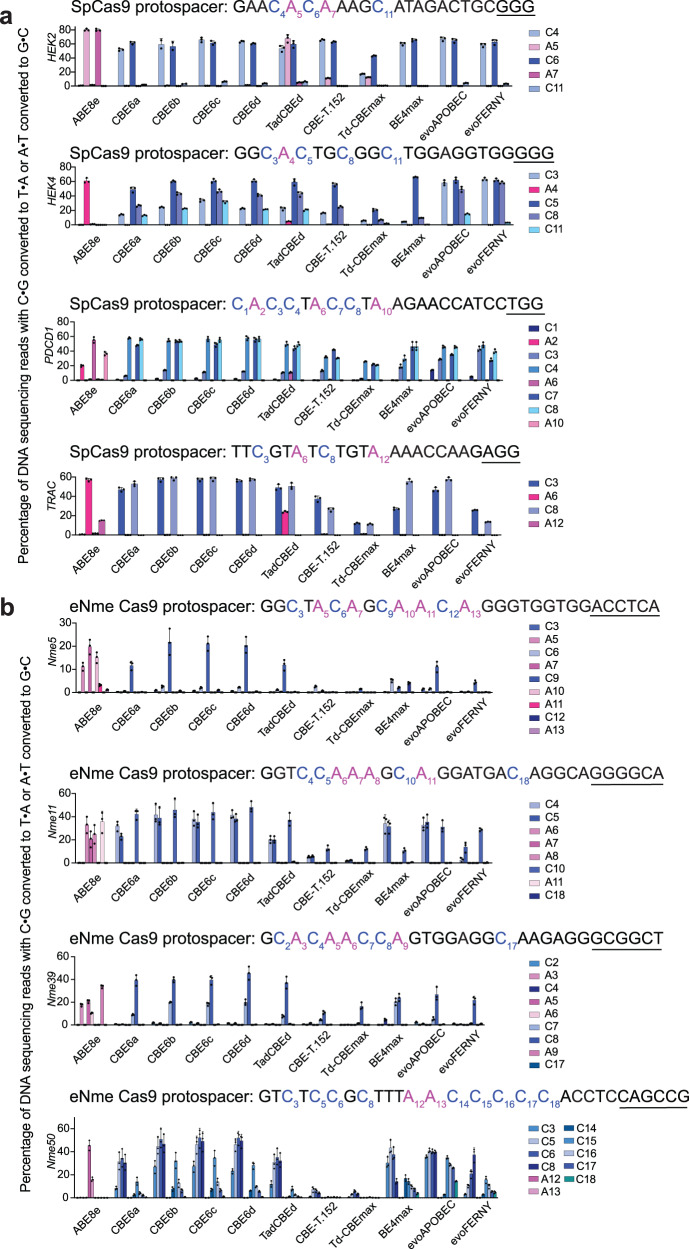
Fig. 3. Comparison of CBE6 variants with existing CBEs in mammalian cells.
a CBE6 variants or existing cytosine base editors, all using SpCas9 nickase domains in the BE4max architecture, were transfected into HEK293T cells with guide RNAs targeting three protospacers. Data are presented as mean values ± SD. Dots represent individual values from n = 3 independent biological replicates. PAM sequences are underlined. HEK293T site 2 is abbreviated HEK2, and HEK293T site 4 is abbreviated HEK4. b CBE6 variants along with existing cytosine base editors using eNme2-C Cas9 nickases in the BE4max architecture were transfected into HEK293T cells with guide RNAs targeting three protospacers. Data are presented as mean values ± SD. Dots represent individual values from n = 3 independent biological replicates. PAM sequences are underlined. Full data for C•G-to-T•A and A•T-to-G•C activity are in Supplementary Figs. 12 and 13. Source data are provided as a Source Data file.