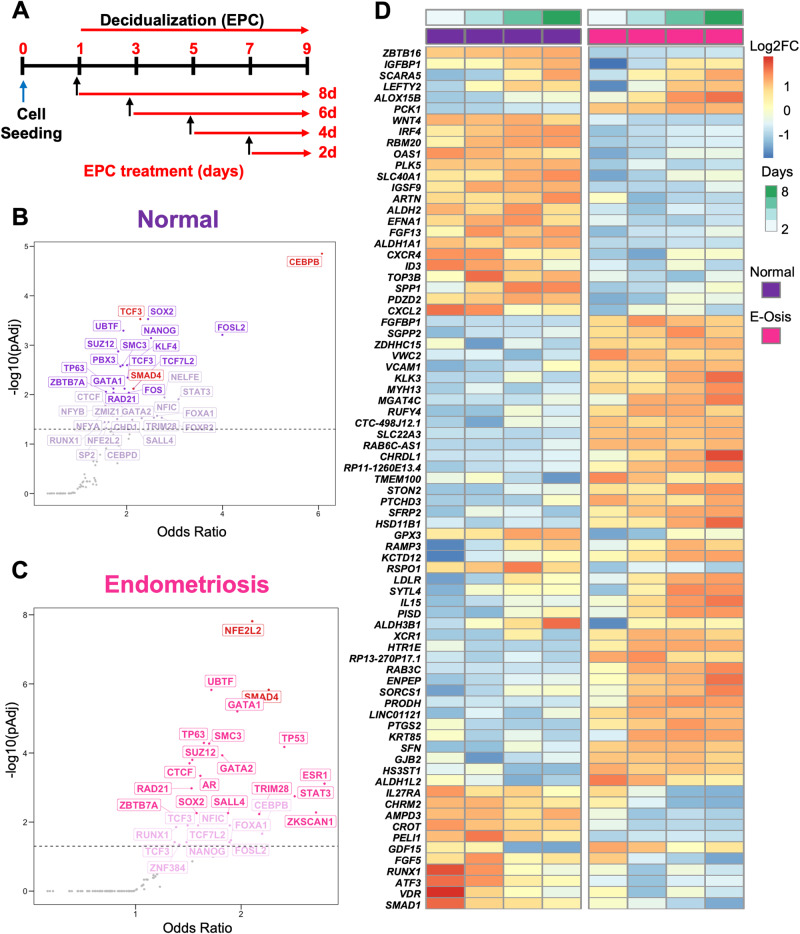
Fig. 1. Transcriptomic profiling of endometrial stromal cells from individuals with or without endometriosis reveals key differences during in vitro decidualization.
A Primary endometrial stromal cell cultures from individuals without (n = 3, “normal”) or with endometriosis (n = 4) were subjected to a time-course decidualization treatment. After plating, cells were treated with vehicle or with the decidualization cocktail (35 nM estradiol, 1 µM medroxyprogesterone acetate, 50 µM cAMP, “EPC”) for 2, 4, 6, or 8 days. RNA sequencing was performed and the decidualization response within normal and endometriosis stromal cells was determined by normalizing differentially expressed genes relative to the Day 0 (vehicle)-treated cells. B, C Upstream transcriptional regulators were identified by searching for conserved ENCODE and ChEA consensus gene targets among the differentially expressed genes in the normal (B) and endometriosis (C) groups. CEBP/β and TCF3 emerged as top transcription regulators for normal decidualizing cells (B), while NFE2L2 and SMAD4 were determined to be major upstream regulators for endometriosis. D Heatmap displays gene expression over time within the normal and endometriosis (“E-Osis”) groups treated with EPC using normalized z-scores. Color represents log2 fold-change relative to baseline (day 0).