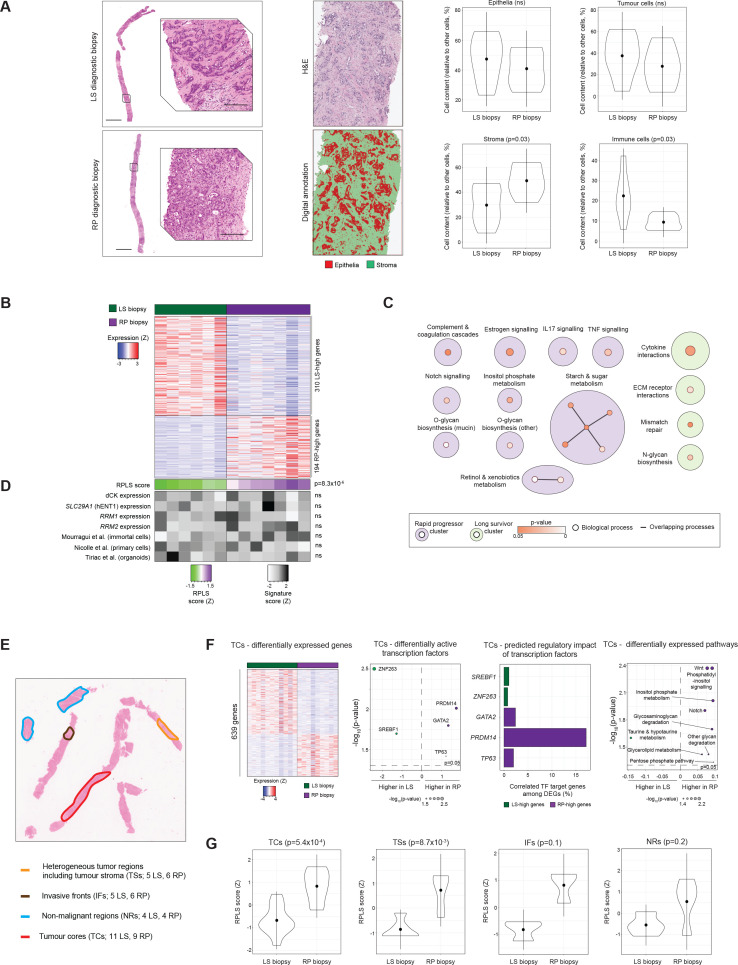
Figure 2.
Histopathological and transcriptomic profiles of diagnostic biopsies from the RPLS cohort. (A) Representative H&E images (scale bars: 2 mm—top; 200 µm—bottom), characterisation of the epithelial component of diagnostic biopsies in each region of interest, and differential composition in tumourous, stromal and immune cells following cell segmentation (LS: n=6; RP: n=7; Welch t-test). (B) Heatmap of 504 differentially expressed genes (≥2 fold change, p<0.05; Wilcoxon) between LS and RP biopsies. (C) KEGG pathway over-representation analysis of LS-high and RP-high genes using EnrichmentMap. Overlapping pathways are connected by lines and annotated under a common theme using AutoAnnotate. KEGG: Kyoto Encyclopaedia of Genes and Genomes. (D) Heatmap and differential expression analysis of the RPLS score and previously published metrics of gemcitabine sensitivity in the RPLS cohort. P values were derived by Wilcoxon test. (E) Representative H&E stain of a diagnostic biopsy, indicating histological regions targeted by macrodissection. (F) Differentially expressed genes (≥2 fold change, p<0.05; Wilcoxon), differentially active transcription factors (p<0.05, Wilcoxon test; DoRothEA), differentially expressed pathways (p<0.05, Wilcoxon test; ssGSEA of KEGG and Hallmarks gene lists), and differentially active cytokines (p<0.05, Wilcoxon test; CytoSig) between RP and LS tumour cores (TCs; 11 LS, 9 RP). (G) Differential expression of the bulk tissue RPLS signature in TCs, tumour stroma (TSs; 5 LS, 6 RP), invasive fronts (IFs; 3 LS, 3 RP) and non-malignant regions (NRs; 4 LS, 4 RP) from RP and LS biopsies (Wilcoxon test). LS, long survivor; RP, rapid progressor.