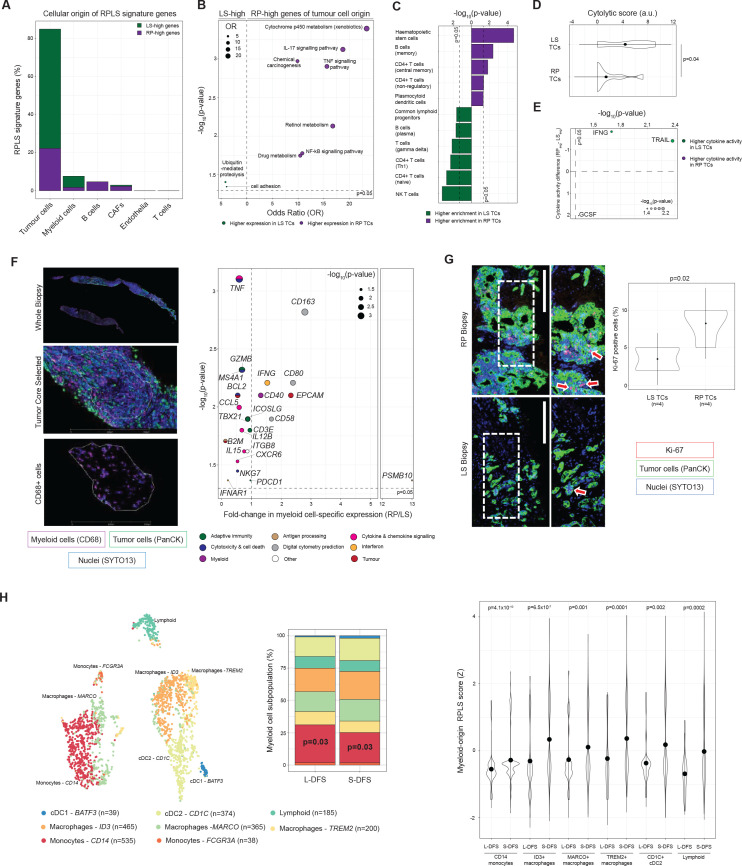
Figure 3.
Cellular origins and tumour-immune dynamics associated with the RPLS signature. (A) Barplot of cell type-associations of RPLS signature genes. Genes were only assigned to cell types if their associations were supported by two independent single cell RNA-sequencing datasets. (B) KEGG pathway over-representation analysis of tumour-origin RPLS signature genes. (C) Differential enrichment (Wilcoxon test) of immune cell type signatures in rapid progressor (RP) and long survivor (LS) tumour cores (TCs) determined by cellular deconvolution (xCell). (D) Differential cytolytic scores (Wilcoxon test) between RP and LS TCs. (E) Differential cytokine activities (Wilcoxon test) between RP and LS TCs determined by CytoSig. (F) Representative multiplex immunofluorescence images of TCs undergoing RNA extraction from myeloid (CD68+) myeloid cells using the Digital Spatial Profiling (GeoMx) platform with regions of interest identifed. Volcano plot of differentially expressed genes (Immune Pathways Panel (NanoString) plus 5 custom targets derived from digital cytometry) in tumour-infiltrating myeloid cells from LS (n=6) and RP (n=6) TCs. P values were computed by Wilcoxon test. (G) Representative Ki-67 staining in an LS and RP TCs, including differential proliferation analysis (Welch t-test). (H) tSNE plot of myeloid subpopulations identified in immune-enriched single cell RNA-sequencing data from three resected iCCA. Frequency barplot (p values from χ2 test) comparing the abundance of myeloid subpopulations in patients without (long disease-free survival, L-DFS) and with (short disease-free survival, S-DFS) recurrence under adjuvant treatment with capecitabine. Differential expression (Wilcoxon test) of the myeloid-origin RPLS signature in myeloid subpopulations. iCCA, intrahepatic cholangiocarcinoma.