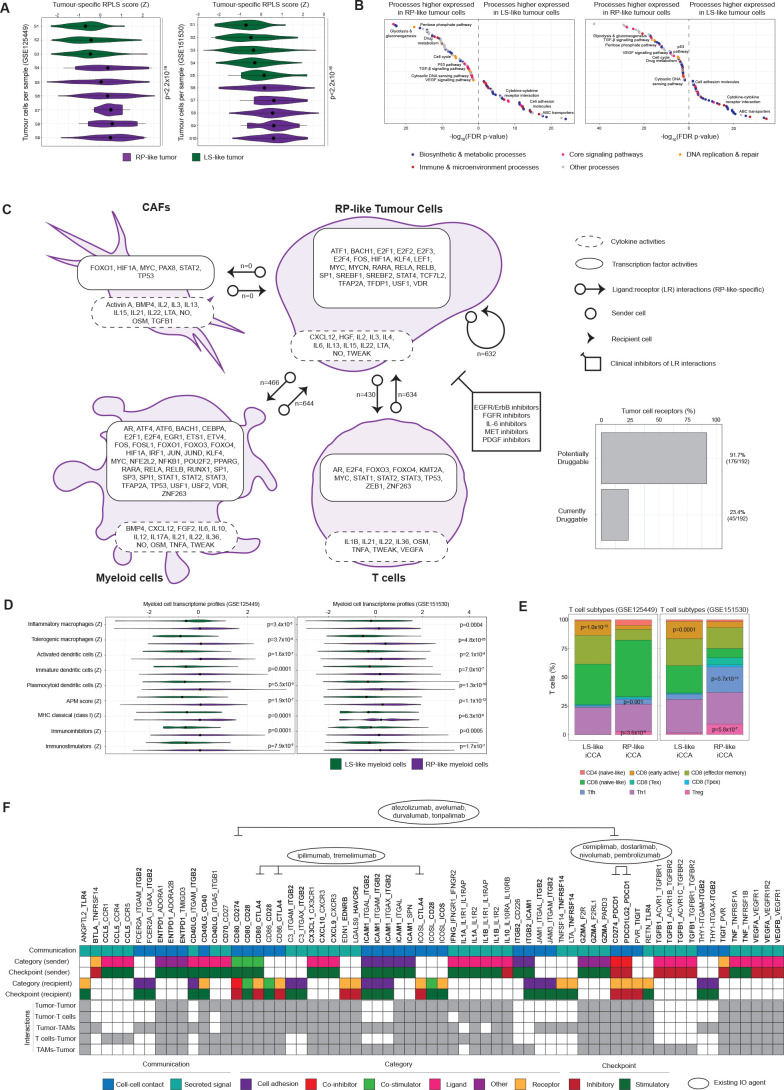
Figure 4.
Modelling the RPLS signature in iCCA single cell RNA-sequencing data. (A) Annotation of tumours as long survivor (LS)-iike or rapid progressor (RP)-like based on tumour cell expression of the tumour-origin RPLS signature (ESCAPE tool, p values derived by Wilcoxon test). (B) Differentially expressed pathways and processes (ESCAPE with KEGG and Hallmarks gene lists) between LS-like and RP-like tumour cells. (C) Cell type-specific transcription factor activities (DoRothEA), cytokine activities (CytoSig) and ligand:receptor interactions (CellChat) unique to RP-like tumours in GSE125449 and GSE151530, including the potential and current druggability of tumour surface receptors. (D) Differential expression of myeloid cell type and functional signatures in LS-like and RP-like myeloid cells (ESCAPE, Wilcoxon test). (E) T cell subtype annotation using ProjectTILs (p values from Fisher’s exact test). (F) RP-specific ligand-receptor interactions tumours in GSE125449 and GSE151530 involving immunomodulatory targets (highlighted in bold; defined by CRI iAtlas). iCCA, intrahepatic cholangiocarcinoma.