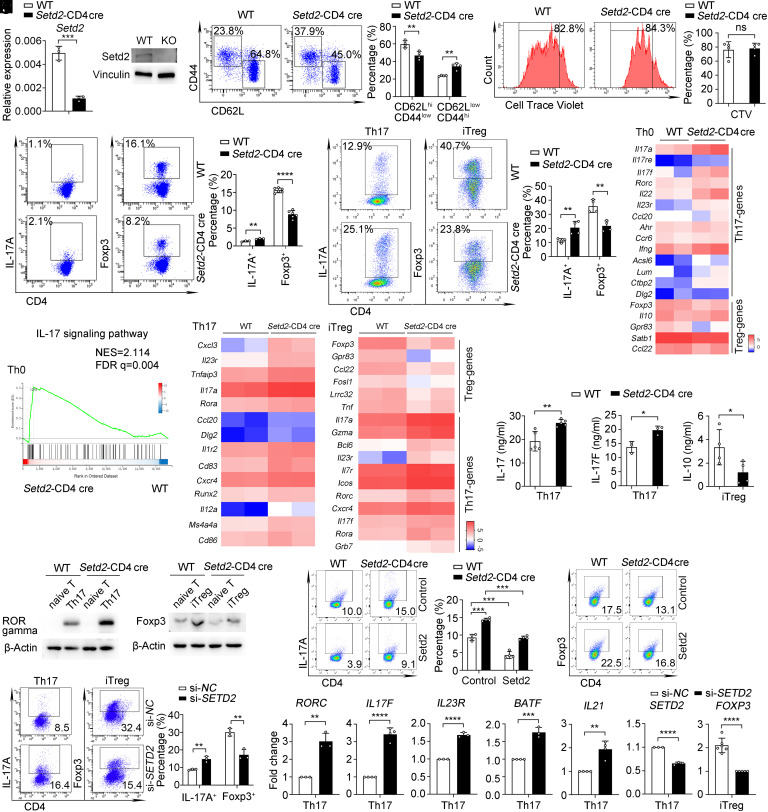
Fig. 2.
Deletion of Setd2 promotes Th17 but suppresses Treg cell polarization. (A) mRNA and protein levels of Setd2 expression in CD4+ T cells of wild-type (WT) and Setd2-CD4 cre mice (n = 3). (B) Flow cytometric analysis of CD62L+ and CD44+ in splenic CD4+ T cells from WT and Setd2-CD4 cre mice (n = 4). (C) CD4+ T cells proliferation after treatment with αCD3 (5 μg/mL) and αCD28 (2 μg/mL) for 72 h (n = 4). (D and E) Flow cytometric analysis and quantification of IL-17A+ and Foxp3+ cells in splenic CD4+ T cells (D) and differentiated Th17 and iTreg cells for 3 d (E) (n = 3 to 6). (F) Heatmap shows the differential genes from Th0 cells (fold change > 1.5, P < 0.05). (G) GSEA analysis of enrichment in IL-17 signaling pathway-related genes from Th0 cells. (H) Heatmap showing genes differentially expressed in Th17 cells and iTreg cells (fold change ≥ 2, P < 0.05). (I) CBA analysis of IL-17, IL-17F, and IL-10 in Th17 or iTreg cell supernatants (n = 3 to 5). (J) Immunoblot analysis of RORγt and Foxp3 levels in Th17 and iTreg cells. (K and L) Flow cytometric analysis of IL-17A+ and Foxp3+ cells transfected with Setd2 expressing lentivirus (K) and SETD2 siRNA (si-SETD2) or control siRNA (si-NC) (L) in Th17 and iTreg cells (n = 3 to 4). (M) qPCR analysis of Th17 and Treg-genes expression in human Th17 or iTreg cells transfected with si-SETD2 or si-NC (n = 3 to 6). Data show one of three independent experiments. Results are presented as means ± SD (A–E, I, and K–M). *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001, and ns, not significant. FDR, false discovery rate. NES, normalized enrichment scores.