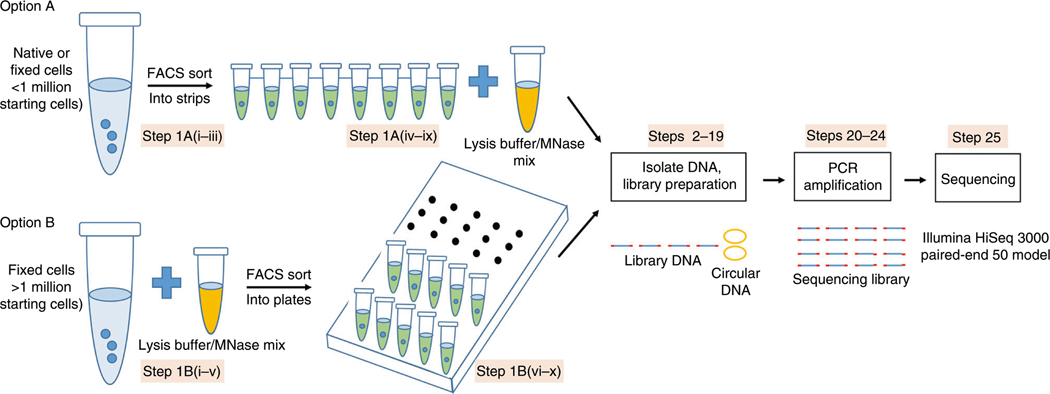
Fig. 1 |. Workflow illustration of the scMNase-seq protocol.
There are two options for generating scMNase libraries according to the starting cell number. For <1 million starting cells, single cells are isolated first by FACS sorting, and then each cell is individually digested by MNase digestion. For >1 million starting cells, bulk cells are fixed and digested by MNase first, and then single cells are isolated by FACS sorting. Both options follow the same DNA extraction procedure by phenol–chloroform. Circular DNA is spiked-in to prevent DNA loss during processing of the samples. End-repair, A/T ligation and PCR amplification are performed to prepare scMNase-seq libraries. After isolation of desired DNA fragments, the libraries are sequenced on an Illumina sequencing platform.