FIG. 5.
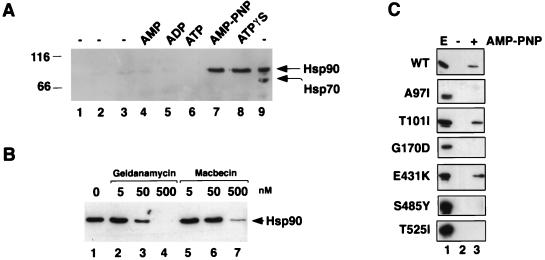
In vitro association of Sba1His6 with Hsp90. (A) Purified Sba1His6 was prebound to Ni-NTA resin and added to desalted whole-cell extracts from wild-type yeast cells and incubated at 30°C for 30 min. Hsp90 binding to Sba1His6 (lanes 1 to 8; lane 9 is whole-cell extract loaded directly onto the gel) was determined after reisolation of Sba1His6 and Western blotting using anti-Hsp90. Lane 1, Ni-NTA-Sba1His6 incubated in buffer; lane 2, Ni-NTA resin without prebound Sba1His6 incubated with extract; lane 3; Ni-NTA-Sba1His6 incubated with extract but without further addition; lane 4, Ni-NTA-Sba1His6 incubated with extract plus 5 mM AMP; lane 5, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ADP; lane 6, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ATP; lane 7, Ni-NTA-Sba1His6 incubated with extract plus 5 mM AMP-PNP; lane 8, Ni-NTA-Sba1His6 incubated with extract plus 5 mM ATPγS; lane 9, 2.5 μg of desalted whole-cell extract. The blot was first probed with anti-Hsp90 and subsequently reprobed with anti-Hsp70. Molecular size markers are shown in kilodaltons at left. (B) Binding of Hsp90 to Sba1His6 is competed by benzoquinoid ansamycins. Ni-NTA-Sba1His6 resin was incubated with desalted extracts as described above plus 5 mM AMP-PNP in the presence of solvent (dimethyl sulfoxide; lane 1), geldanamycin (lanes 2 to 4), or macbecin (lanes 5 to 7) at the concentrations indicated. After 30 min at 30°C, the resin was reisolated and washed, and bound proteins were resolved by SDS-PAGE. Hsp90 was detected by Western blotting. (C) Binding of wild-type (wt) and different mutant forms of Hsp90 to Ni-NTA-Sba1His6 resin. Lane 1, whole-cell extract (E); lane 2, incubation of Ni-NTA-Sba1His6 resin with extracts in the absence of AMP-PNP; lane 3, incubation of Ni-NTA-Sba1His6 resin with extracts in the presence of AMP-PNP. The Western blots were probed with anti-Hsp90.