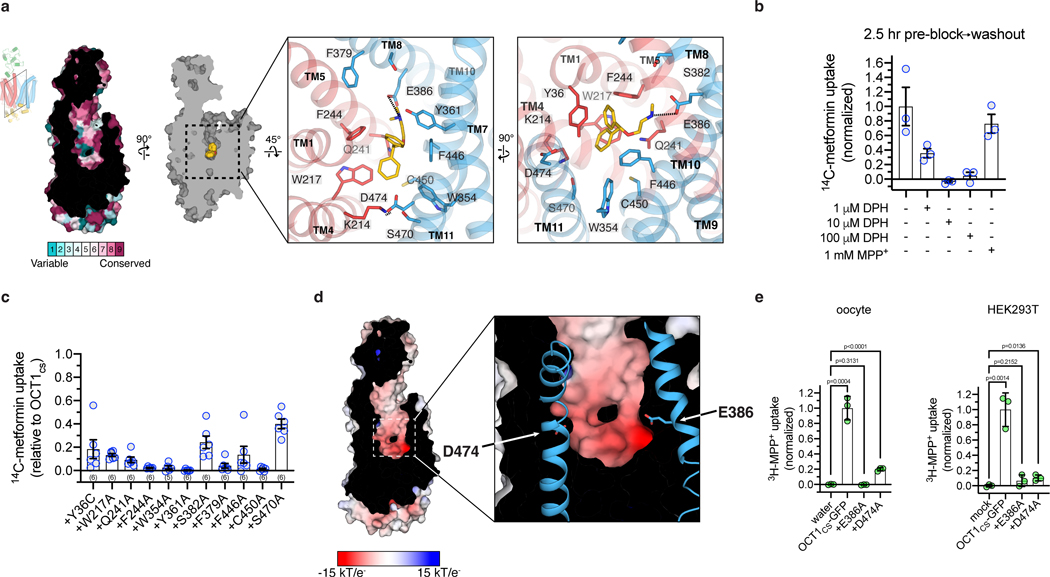
Figure 2 |. Diphenhydramine recognition by OCT1.
a, ConSurf54 analysis of the OCT1CS central cavity (left). Residue Y36 in the central cavity shows high variability across OCT1 orthologs in the multiple sequence alignment. Detailed DPH-OCT1 interactions in the binding cavity (right), highlighting interacting residues. b, Residual cold competition block of 14C-metformin uptake mediated by OCT1CS. 2.5 h pre-treatment with cold competitor performed at the noted concentration, followed by rapid and extensive oocyte washing in ligand-free buffer immediately before initiation of the uptake assay (10 μM 14C-metformin in 30 min into OCT1CS expressing oocytes; data background corrected using the water injected controls, and normalized to none-added control condition; n=3 biologically independent replicates shown along with mean ± s.e.m.). c, Functional evaluation of mutants in the OCT1CS background (accumulation of 10 μM 14C-metformin in 1 h into mutant-expressing oocytes; ; n=3 biologically independent replicates shown along with mean ± s.e.m.). d, APBS55 surface electrostatic calculation of the OCT1CS central cavity (see Methods). e, Left, uptake of 3H-MPP+ by OCT1CS-GFP or mutants in the OCT1CS-GFP background in oocytes (accumulation of 100 nM 3H-MPP+ in 1 h into mutant-expressing oocytes; n=3 biologically independent replicates shown along with mean ± s.e.m. Data background corrected using the water injected condition, and normalized to the OCT1CS-GFP uptake condition). Right, uptake of 3H-MPP+ by OCT1CS-GFP or mutants in the OCT1CS-GFP background expressed in HEK293T cells (accumulation of 10 nM 3H-MPP+ in 40 min into mutant-expressing HEK293T cells; n=3 biologically independent replicates shown along with mean ± s.e.m, data background corrected using the mock-transfected condition and normalized to the OCT1CS-GFP uptake condition). P-values from an unpaired two-tailed t-tests are shown.