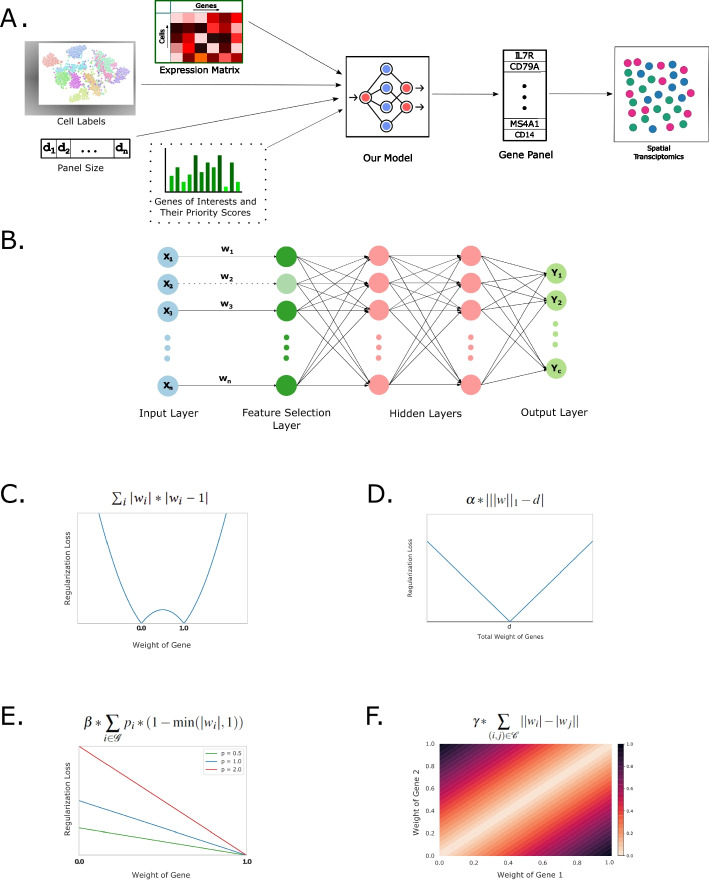
Fig. 1.
Overview of scGIST. A scGIST takes as input expression values from an scRNA-seq experiment, labels for the cells, a target gene panel size, and optionally a list of genes of interest and their priority scores. It uses a deep learning model to learn a set of genes to predict cell types which can be used as a gene panel for single-cell spatial transcriptomics assays. B The neural network model of scGIST consists of a one-to-one feature selection layer connected to the input layer. This is then followed by two fully connected layers and finally an output layer. The model has a custom loss function to C drive weights in the feature selection layer towards 0 or 1, D select the number of genes equal to the panel size (d), E prioritize genes of interest according to user provided priority scores (), and F select both or none of the genes from specified complexes