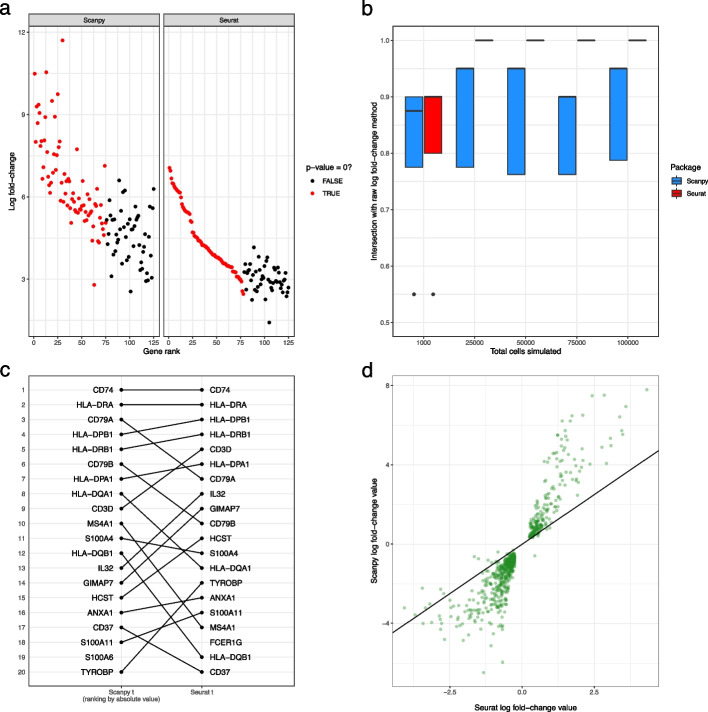
Fig. 7.
Case studies scrutinizing Scanpy and Seurat. a Gene rank vs log fold-change values for the Scanpy Wilcoxon (with tie correction, ranking by the absolute value of the score) and Seurat Wilcoxon methods for the Oligodendrocyte cell type cluster in the Zeisel dataset. The color of the point indicates whether or not the gene has an exactly zero p-value. b The proportion of top 20 genes shared between methods implemented in Scanpy and Seurat and the method of ranking genes by the raw log fold-change calculation on simulated datasets with increasing number of total cells. These simulations have parameters estimated from the pbmc3k dataset and a location parameter for the DE factor of 3. c Visualization of difference in rankings between the Scanpy t method (ranking by the absolute value of the score) and Seurat’s t-test method on the B cell cluster in the pbmc3k dataset. d Scatter plot between the log fold-change values calculated by Seurat and Scanpy on the B cell cluster in the pbmc3k dataset