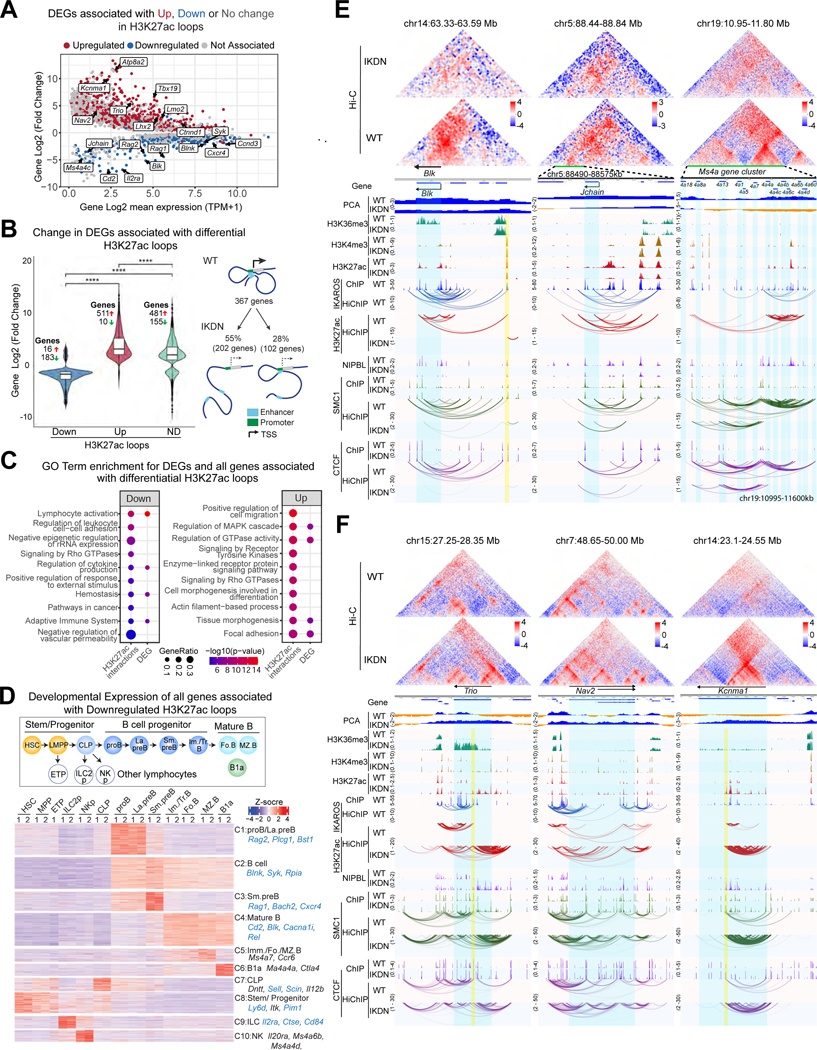
Figure 2. IKAROS-dependent 3D chromatin organization controls B cell identity.
(A) Change in gene expression between WT and IKDN (DEGs) and overlap with H3K27ac HiChIP loops that are up (red), down (blue) or unchanged (grey). (B) DEGs associated with differential (Down and Up) and non-differential (ND) loops identified by H3K27Ac HiChIP. **** p < 0.0001. (C) GO pathway enrichments for all genes associated with differential H3K27ac loops or DEGs. Statistical significance (−log10p-value) is shown by color scale and gene ratio by circle size. (D) K-means clustering of expression during WT hemo-lymphopoiesis and B cell differentiation of all genes associated with downregulated loop anchors. Examples downregulated in IKDN pre-B cells are highlighted in blue. (E) Examples of down-regulated and (F) up-regulated genes with changes in long-distance interactions. Hi-C contact maps (Observed/Expected, 5kb resolution). Compartment affiliation (PCA): euchromatin-blue, heterochromatin-orange. Normalized histone modification and transcription factor ChIP-seq tracks are shown. Interactions identified by FitHiChIP for IKAROS, H3K27ac, SMC1 and CTCF are shown. Highlights: blue -genes, yellow-re-directed loop anchors. Source: Tables S2-4.