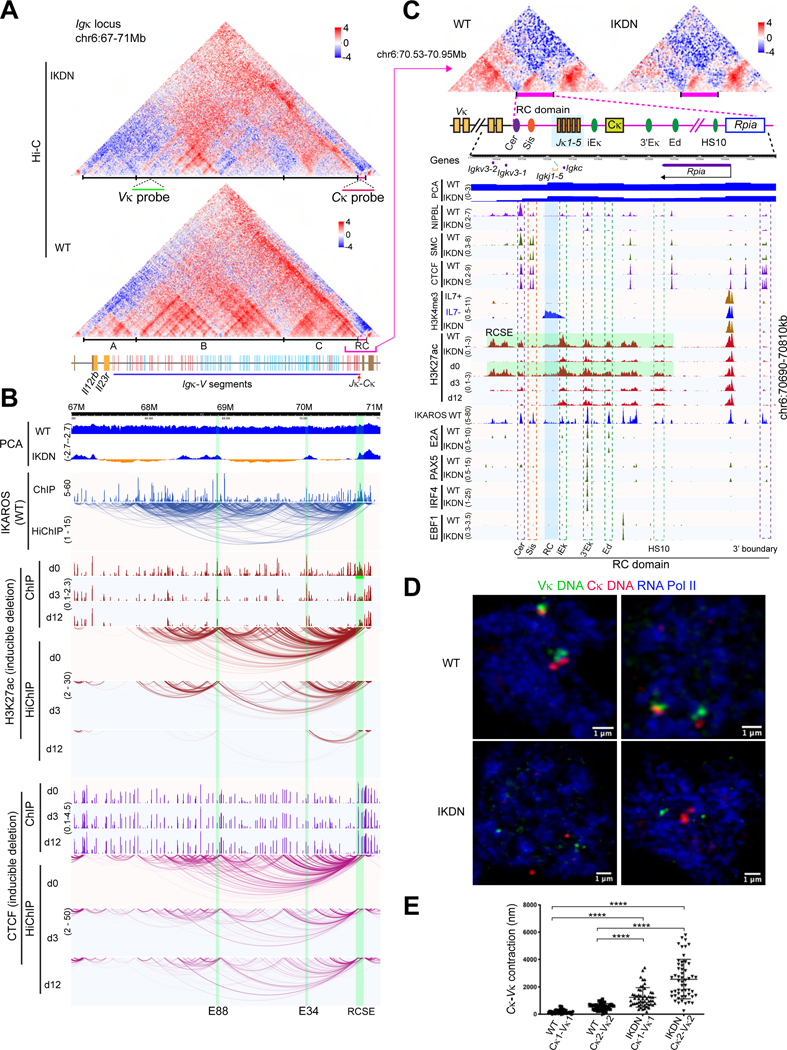
Figure 6. Regulation of chromatin organization and locus contraction at the Igκ locus.
(A) Hi-C at the Igκ locus for WT and IKDN large pre-B cells (10kb). TADs (A-C) and FISH probes are marked. Orientation of Igκ-V segments relative to the J segments is indicated (red-forward and blue-reverse). (B) Genomic tracks as in Figure 5H. (C) The 3’ boundary domain is shown with ChIPseq peaks for indicated chromatin marks, B lineage TFs and other factors. Regulatory (iEκ, 3’Eκ, Ed, HS10) and structural (Cer, Sis) elements and the RCSE (green) that spans them are highlighted. (D) Igκ locus DNA-Immuno FISH using the Vκ (green) and Cκ (red) probes in WT and IKDN pre-B cells (pSer2 RNA Pol II (blue). (E) Distance between Vκ-Cκ regions in WT and IKDN large pre-B cells, **** p < 0.0001). Source: Tables S2-3, S6-7.