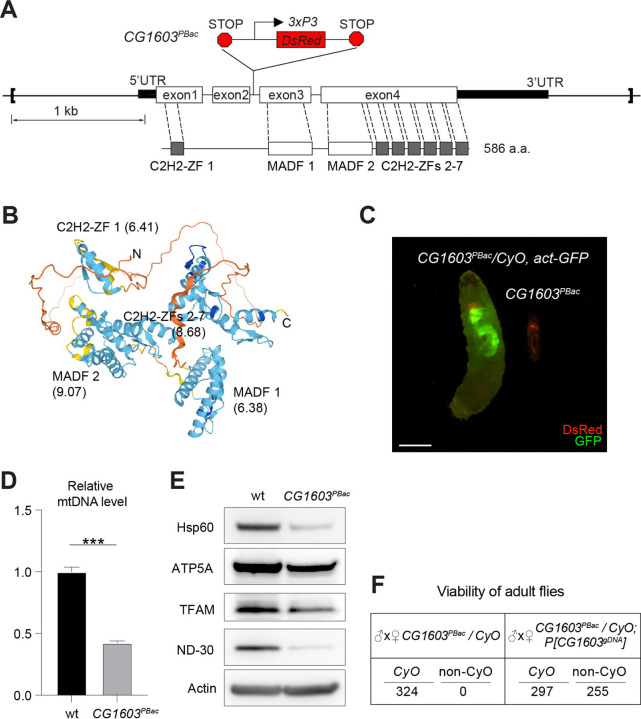
Figure 4. CG1603 gene model, product, mutant and the genomic DNA transgene.
(A) Schematic representation of CG1603 genomic locus, showing the CG1603 transcript (5’ and 3’UTR in black bar and four exons in white), its protein product (586 amino acids in length, and characterized by seven C2H2-ZF and two MADF domains), the CG1603PBac mutant allele (with a piggyBac insertion in the second intron, which is marked by fluorescent DsRed driven by an eye-specific 3xP3 promoter and flanked by stop codons in all three reading frames terminating translation through downstream), and the genomic region (in square brackets, from 955 bp upstream of the CG1603 5’UTR to 656 bp downstream of CG1603 3’UTR) used for P[CG1603gDNA] transgene. (B) Predicted 3D structure of the CG1603 protein by AlphaFold. Labels indicate the N-and C-terminus, as well as the specific protein domains along with their predicted isoelectric point (pI). (C) Images of CG1603PBac / CyO, Act-GFP and homozygous CG1603PBac larvae cultured together at 25°C, day 4 after egg laying. Green: GFP; Red: DsRed. Scale bars: 1 mm. (D) Relative mtDNA levels in CG1603PBac mutant larvae to wild type (wt) control. n= 3, error bar: SD. ***: p<0.001. (E) Western blots of mitochondrial proteins in CG1603PBac mutant larvae to wild type (wt) control. (F) P[CG1603gDNA] restored viability of CG1603PBacflies. The number of progenies for each genotype is listed.