Abstract
Molecular dissection of the B-cell-specific transcription coactivator OCA-B has revealed distinct regions important, respectively, for recruitment to immunoglobulin promoters through interaction with octamer-bound Oct-1 and for subsequent coactivator function. Further analysis of general coactivator requirements showed that selective removal of PC4 from the essential USA fraction severely impairs Oct-1 and OCA-B function in a cell-free system reconstituted with partially purified factors. Full activity can be restored by the combined action of recombinant PC4 and the PC4-depleted USA fraction, thus suggesting a joint requirement for PC4 and another, USA-derived component(s) for optimal function of Oct-1/OCA-B in the reconstituted system. Indeed, USA-derived PC2 was found to act synergistically with PC4 in reproducing the function of intact USA in the assay system. Consistent with the requirement for PC4 in the reconstituted system, OCA-B was found to interact directly with PC4. Surprisingly, however, removal of PC4 from the unfractionated nuclear extract has no detrimental effect on OCA-B/Oct-1-dependent transcription. These results lead to a general model for the synergistic function of activation domains in Oct-1 and OCA-B (mediated by the combined action of the multiple USA components) and, further, suggest a functional redundancy in general coactivators.
The B-cell-specific function of immunoglobulin (Ig) promoters is mediated by an octamer element (ATTTGCAT) and was thought originally to be regulated by an octamer-binding factor (Oct-2) enriched in lymphoid cells (reviewed in reference 51). However, early biochemical analyses indicated that Ig promoters could function equally well with Oct-2 or the ubiquitous Oct-1 and that B-cell-specific promoter function was determined by a B-cell-specific Oct-1-associated coactivator designated OCA-B (30, 41). These studies established a new paradigm for cell-specific promoter activation that involves recruitment of the true regulatory factor to the cell-specific promoter element through interaction with a ubiquitous DNA binding factor. Subsequent genetic analyses confirmed that Oct-2 was nonessential for Ig promoter activation but failed to identify the responsible B-cell-specific factor (6–8).
The discovery of OCA-B set the stage for cloning of a cognate cDNA on the basis of biochemical purification (31) and yeast genetic screens (13, 54), which, in turn, facilitated both genetic and biochemical analyses of OCA-B. Targeted gene disruption data (19, 39, 50) revealed that OCA-B is essential for normal patterns of Ig expression, most notably antigen-dependent responses leading to secondary isotype production, but not for antigen-independent Ig gene transcription events that might conceivably employ other Oct-1 coactivators.
Biochemical studies with recombinant OCA-B have confirmed the originally suggested mechanisms by showing that the highly related POU domains (reviewed in references 15, 16, and 45–48) of Oct-1 and Oct-2, but not that of Oct-3, are sufficient for OCA-B interaction and promoter recruitment (13, 31, 54). More recent studies have shown that OCA-B also contacts octamer nucleotides through the major groove in the DNA–Oct-1–OCA-B complex (2, 4), consistent with a stronger OCA-B interaction with Oct-1 or Oct-2 in the presence of DNA (31). Further, the demonstration that OCA-B can discriminate among octamer variants for formation of the higher-order ternary complex provided a mechanism for differential activation of octamer-containing promoters (4, 14), whereas the inability of OCA-B to stimulate the histone 2B (H2B) promoter (31), which contains a functional octamer element identical to the consensus Ig promoter element, requires a different explanation. Finally, while corresponding POU domains are sufficient for OCA-B recruitment to Ig promoters and for normal octamer-mediated transcription from the H2B promoter, an additional Oct-1 or Oct-2 activation domain(s) is necessary for functional synergy with OCA-B and corresponding activation of Ig promoters (31). When assayed in more purified reconstituted system, Ig promoter activation by OCA-B and Oct-1 also required the general coactivator fraction USA in addition to the general initiation factors (31).
The availability of recombinant OCA-B and in vitro assay systems with general initiation factors (reviewed in reference 44) and cofactors (reviewed in reference 18) prompt questions about structure-function relationships in relation to its interaction both with Oct-1 (upstream interactions) and, potentially, with components of the general transcription machinery (downstream interactions). As predicted from our previous model invoking separate domains for these interactions (31), mutant OCA-B defective in either interaction would, in principle, lead to defects in coactivation function. This scenario is reminiscent of that described for the herpes simplex virus (HSV) (co)activator VP16, which was reported to interact with promoter-bound Oct-1 through the POU domain (e.g., 26, 43); have a conditional DNA binding activity (25, 52, but see reference 58; reviewed in references 5 and 15), a property shared by OCA-B (2, 4); and provide an activation function. From this point of view, OCA-B may represent a class of cellular counterparts of viral coactivators (see Discussion).
In this report, we first show functional analyses of wild-type OCA-B versus mutant derivatives to define domains critical for upstream versus downstream interactions. We then present a further definition of general cofactor requirements for the proper function of Oct-1 and OCA-B in a reconstituted system. Finally, we use OCA-B mutants to establish a functional linkage between OCA-B and general cofactors.
MATERIALS AND METHODS
In vitro transcription.
For transcription in nuclear extracts, conditions described by Luo et al. (30) and Luo and Roeder (31) were used. The reaction mixtures (25 μl) contained 5 μl of transcription premix (200 mM HEPES [pH 8.4]; 15 mM MgCl2, 3 mM each ATP, GTP, CTP, and UTP; 20 mM dithiothreitol, 40 U of RNasin); 5 μl of H2O containing 500 ng of the BCL1 (27) IgH template (41) and, when indicated, 50 ng of the 2×Sp1 template (42); and 15 μl of a mixture of nuclear extract maintained in BC100 (30; typically, 10 μl) and various amounts of cofactor OCA-B maintained in BC100–250-μg/ml bovine serum albumin (BSA) with compensating volumes of the same buffer. For transcription in a reconstituted system, the nuclear extract–OCA-B mixture mentioned above was replaced with TFII-A, -B, -D, and -E/F/H plus RNA polymerase II and other factor and cofactor components specified in the figures and legends. Factors in this system were stabilized by 1.5-mg/ml BSA. All of the in vitro transcription reaction mixtures were incubated at 30°C for 60 min, and transcripts were measured by primer extension.
Purification of factors and cofactors.
Proteins were maintained in the BC buffers described by Luo et al. (30). Numbers following BC indicate millimolar KCl concentrations. Highly purified factors and cofactors (in BC100) were stabilized with 250-μg/ml BSA. Oct-1 (glycosylated) was purified from HeLa nuclear extracts by using a combination of wheat germ agglutinin and octamer binding site affinity columns as described by Pierani et al. (41). General transcription factors and cofactors were prepared as described by Ge et al. (10) and Kretzschmar et al. (24).
Recombinant wild-type and mutant OCA-B proteins were expressed as six-His-tagged proteins (vector 6His-pET11d; reference 17) in Escherichia coli and purified. Recombinant PC4 was expressed and purified as described by Ge et al. (10).
Immunodepletion of PC4.
Immunodepletion of PC4 was carried out at 4°C. The USA fraction from the heparin-Sepharose step (see reference 34; 0.4 ml in BC100, diluted to 2 ml with BC600) was passed several times through a 0.4-ml protein A-agarose column equilibrated in BC500. Half of the flowthrough fraction served as a source of mock-depleted USA. The other half was passed several times through a 0.2-ml anti-PC4 affinity column equilibrated in BC500. Both the final flowthrough fraction from the protein A column and that from the anti-PC4 column were dialyzed to BC100, concentrated (about fivefold) by ultrafiltration (Amicon, Inc.), and used as mock-depleted and PC4-depleted USA fractions in transcription assays. The extent of depletion was monitored by immunoblotting various fractions with anti-PC4 sera (see Fig. 3A). Removal of PC4 from HeLa nuclear extract was carried out by a similar protocol, except for the omission of the initial dilution and final concentration steps.
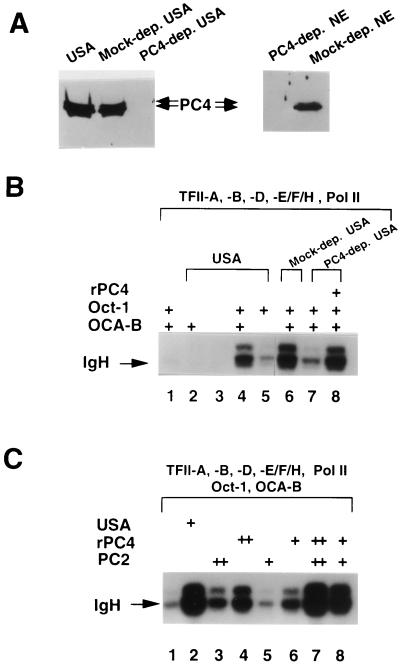
FIG. 3.
PC4 as an essential component of USA and acting synergistically with PC2, to support the function of OCA-B in a reconstituted transcription system. (A) PC4 depletion (dep.) from the USA coactivator fraction (left panel) and from the nuclear extract (NE; right panel; the treated nuclear extracts were used in the experiment shown in Fig. 5). The completion of the depletion was examined by immunoblotting with anti-PC4 antibodies. (B) IgH promoter activity was analyzed in the reconstituted system with components of general factors and RNA polymerase II (top) in the presence (+) or absence of Oct-1, OCA-B, or the USA fraction, as indicated. Lanes 1 to 5 demonstrate an IgH promoter dependency on activator Oct-1, general coactivator fraction USA, and specialized coactivator OCA-B. Oct-1 is absolutely required for the promoter activity (compare lanes 2 and 3 to lane 4, which represents a complete set of components sufficient to bring about a high level of IgH promoter transcription). In an otherwise complete system, the transcription is stimulated ∼15-fold by USA and ∼8-fold by OCA-B (compare lane 1, missing USA, and lane 5, missing OCA-B, respectively, to lane 4, complete). Lanes 6 to 8 demonstrate that PC4 is an essential component of USA to support the function of OCA-B in the reconstituted transcription system. The defect of the PC4-depleted USA fraction (lane 7), compared to the complete USA fraction (lane 6), can be rescued by the addition of 50 ng of recombinant PC4 (lane 8). (C) PC4-PC2 synergism for the function of OCA-B in the reconstituted system. Components (top) were used to transcribe the IgH promoter in the absence or presence (+ or ++) of a general coactivator(s), as indicated. The levels of coactivation are (compared to lane 1) ∼15-fold by USA (lane 2), ∼1.5- to 3-fold by PC2 (lanes 5 and 3), ∼3- to 5-fold by PC4 (lanes 6 and 4), and ∼10- to 15-fold by PC2 plus PC4 (lanes 8 and 7). Amounts of factors used in the reconstituted system (for both B and C) are as follows: TFII-A, 0.5 μl; recombinant TFII-B, 50 ng; TFII-D, 2 μl; TFII-E/F/H fraction, 3.5 μl; RNA polymerase II, 0.25 μl; Oct-1 (when added), 20 ng; recombinant OCA-B, 50 ng; recombinant PC4, 25 (+) and 50 (++) ng; USA, mock-depleted USA and PC4-depleted USA, 1 μl of each; PC2, 1 (+) and 2 (++) μl.
Anti-PC4 antibodies were raised against a protein containing full-length PC4 fused to glutathione S-transferase (GST). Specific anti-PC4 antibodies were purified from crude antisera by using an immunogen affinity column. The purified antibodies were then cross-linked to protein A-agarose, yielding an anti-PC4 affinity column.
Protein-protein interaction assays.
To detect a protein(s) that can specifically bind to the wild-type OCA-B activation domain, GST and a GST–OCA-B fusion protein (containing residues 171 to 256 in wild-type and mutant B OCA-B; Fig. 1) were expressed in E. coli and then purified by binding to glutathione-agarose beads. Subsequently, aliquots of HeLa nuclear extract in BC100 were passed through the resultant beads equilibrated in BC100. After extensive washing of the resins with BC100, the bound proteins were eluted by BC1000. The eluates were analyzed by multiple immunoblots using available antisera against various general factors and cofactors; PC4 was the sole polypeptide detected by immunoblotting.
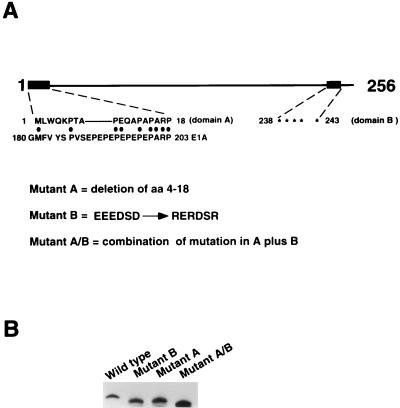
FIG. 1.
(A) A simple linear description of the OCA-B molecule. The 256-residue polypeptide is rich in proline (∼16%; for the complete sequence, see references 13, 31, and 54). Relevant putative functional domains include domain A (with its sequence alignment with an E1A segment shown) and domain B (with acidic residues marked by asterisks). The mutant forms of OCA-B used in this study are also described. The mutant proteins were made by a PCR-based strategy. (B) Protein profile of bacterially expressed, 6His-tagged wild-type OCA-B and mutant forms of OCA-B. A roughly calculated 0.1 ng each of the proteins was analyzed by immunoblotting with polyclonal anti-OCA-B antibodies. (Given that all three mutant proteins may have lost some epitopes compared to the wild type, the amounts of the mutant proteins might be slightly underestimated. Nevertheless, this underestimation would not complicate our analyses of, and the conclusions made about, these mutant proteins.) The amounts used for the in vitro transcription analyses in Fig. 2 were based on this estimation.
Analysis of the in vivo function of OCA-B and derived mutants by transfection.
cDNAs encoding wild-type and mutant (A, B, and A/B) OCA-B proteins were subcloned into mammalian expression vector pRC (Invitrogen). These effector constructs were cotransfected with a reporter gene (luciferase in vector pGL3; Promega) under the control of the BCL1 (27) IgH promoter. The transfection analyses were carried out with HeLa cells by using conventional calcium phosphate methods, and cells were harvested after 48 h. Extracts were prepared by using reporter lysis buffer (Promega), and expression was assayed in accordance with the manufacturer’s instructions. In each case, assays were conducted in duplicate or triplicate and a constant amount of the vector CMVβgal was added as an internal control for transfection efficiency. The expression vector levels were titrated carefully, and the amounts used for the data included here were within the linear range for activation. Western blot analyses indicated that the OCA-B cDNA and the various mutant forms were expressed at similar levels (data not shown).
RESULTS
Molecular dissection of OCA-B.
In addition to being rich in proline (∼16%), a characteristic of a number of transcription factors (reviewed in reference 35), OCA-B displays a weak sequence similarity to the viral coactivator E1A (31; domain A in Fig. 1A) and contains a putative acidic activation domain (domain B in Fig. 1A) reminiscent of that of coactivator VP16 (56). To explore the possibility that these domains are important for OCA-B function, three OCA-B mutants were made as described in the legend to Fig. 1. The A mutant protein has domain A (amino acids [aa] 4 to 18) deleted, the B mutant protein has the acidic stretch neutralized, and the A/B mutant protein is a combinatorial double mutant protein. For in vitro analyses, wild-type OCA-B and the mutant forms of OCA-B were expressed in and purified from bacteria (Fig. 1B); they then were tested for the ability to stimulate an Ig heavy-chain (IgH, BCL1 [27rsqb;) reporter promoter in a HeLa cell nuclear extract. In a dose-response experiment (Fig. 2A), all three mutant proteins showed reduced transcriptional activity compared to wild-type OCA-B. To analyze in vivo functions, HeLa cells were cotransfected with the BCL1 IgH promoter reporter and cytomegalovirus-driven expression vectors. As expected, the in vivo activities of the wild-type and mutant OCA-B proteins on the IgH promoter paralleled those observed in vitro (Fig. 2B). Both in vivo (Fig. 2B) and in vitro (Fig. 2A), the defect in the combinatorial A/B mutant protein was more severe than that of either the A or the B mutant protein. Quantitation of the activation levels is indicated in the figure legends.
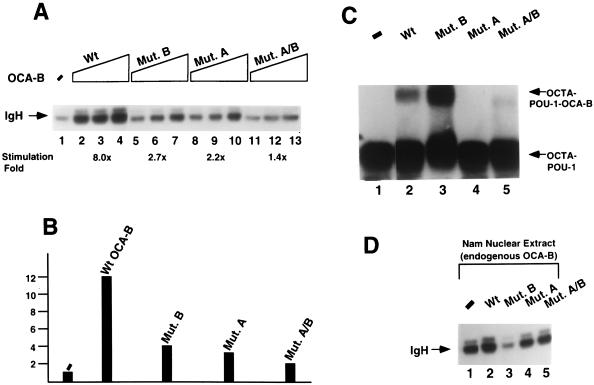
FIG. 2.
Analyses of wild-type (Wt) OCA-B and mutant (Mut.) forms of OCA-B. (A) In vitro transcription assay. As indicated, three doses of wild-type OCA-B and mutant OCA-B (respectively, 10 ng [lanes 2, 5, 8, and 11]; 20 ng [lanes 3, 6, 9, and 12], and 50 ng [lanes 4, 7, 10, and 13]) were used in the titration experiment to complement a HeLa nuclear extract (10 μl for each reaction). The transcripts were analyzed by primer extension as described by Luo and Roeder (31). Lane 1 (−) was a control (without the addition of OCA-B or its derivatives). Transcription activation levels (by the middle point dosage of OCA-B, i.e., 20 ng; an amount equivalent to that of the endogenous OCA-B present in 10 μl of the B-cell [Nam] nuclear extract), compared to lane 1 (taken as 1), are 8, 2.7, 2.2, and 1.4, respectively, for the wild type and the B, A, and A/B mutant proteins (lanes 3, 6, 9, and 12; also, see the fold stimulation shown under each corresponding lane). (B) In vivo (transfection) assay. An IgH reporter promoter construct (with the promoter inserted in front of the luciferase gene) was cotransfected with effector constructs (expressing OCA-B or its mutants under the control of the cytomegalovirus promoter) in HeLa cells. Relative IgH promoter activation levels were scored by measuring the light units of the extracts of the transfected cells 48 h posttransfection normalized to β-galactosidase controls. Taking the level with no effector as 1, the relative levels are 12, 4, 3, and 2, respectively, for the wild type and the B, A, and A/B mutant proteins. The mutant OCA-B proteins were expressed at levels similar to that of the wild-type protein in transfected cells, as assessed by immunoblotting with anti-OCA-B antibodies (data not shown). (C) Octamer DNA–POU-1 complex supershift assay. POU-1 is a truncated version of Oct-1 containing only the POU domain, sufficient to bind OCA-B either in solution or when on DNA (e.g., reference 31). The supershift condition was the same as that described by Luo and Roeder (31), except that approximately 25-fold less OCA-B (or its mutants) was used, such that only a portion of the binary complex was supershifted. To obtain the highest possible resolution, the free probe (a labeled DNA fragment containing the IgH octamer site) was allowed to run out of the gel. As can be seen, the B mutant protein (lane 3) retained the ability to form the higher-order complex, which was even denser than that formed with the wild type (lane 2) for a reason we do not know, whereas the A mutant protein (lane 4) lost the capacity to bind the binary complex. Given that the combinatorial A/B mutant protein (lane 5) could give rise to the formation of a residual level of the ternary complex, it is possible that the introduction of the B mutant protein created a conformational change that can increase the POU–OCA-B interaction even with domain A deleted. We emphasize that such a complication in the binding assay did not complicate our in vitro transcription analyses and conclusions. (D) Test of dominant negative potentials of wild-type OCA-B and its mutant forms. Eight microliters of B-cell (Nam) nuclear extract was used for each reaction (there is ∼2 ng of endogenous OCA-B per μl in this nuclear extract [see above]). The nuclear extract was supplemented with either BC100 buffer (lane 1) or, as indicated, recombinant wild-type or mutant OCA-B (∼100 ng, for a molar ratio of ∼6:1 over the endogenous OCA-B) in BC100 (lanes 2 to 5). The transcripts were analyzed as described for panel A.
These mutant B results suggest that domain B is a major activation domain of the OCA-B molecule. Further confirming this notion is the fact that while a Gal4 (1-94)–OCA-B fusion protein functions as an activator on promoters containing Gal4 binding sites, a mutant protein with a deletion encompassing domain B loses the activation capability (53). However, our mutant B results differ somewhat from those reported by Gstaiger et al. (14), who found that large C-terminal deletions (aa 193 to 256 and 123 to 256) had no detrimental effects on the coactivation function of OCA-B in transfection assays. Furthermore, in our transfection assays, even a smaller C-terminal deletion in OCA-B (aa 209 to 256; a deletion encompassing domain B) was found to impair the coactivation function of OCA-B (53). One explanation for this apparent discrepancy is that the more extensive C-terminal deletion mutant proteins of Gstaiger et al., which still had activation function in their assay, may have also lost an intrinsic inhibitory domain that normally masks an otherwise silent activation function (residing between residues 66 and 122). Alternatively, it is possible that in the transfection assays of Gstaiger et al., the effectors were overexpressed, amplifying minor activation domains whose effects normally would not be observed. In agreement with the second possibility, similar mutant proteins, when carefully titrated, were, in fact, defective in our studies (53). Artificial coactivation by OCA-B or its derivatives (when in excess) may also result from their less functionally relevant interactions with DNA or with the basal machinery. Thus, high doses of the A and B mutant proteins and, to a lesser extent, the A/B mutant protein cause detectable stimulation in vitro and in vivo (53). This is exemplified in Fig. 2A, which shows that the largest amounts of mutant OCA-B had measurable effects.
The wild-type and mutant OCA-B proteins were next checked for the ability to supershift a complex containing an octamer element and the DNA binding (POU) domain of Oct-1 in an electrophoretic mobility shift assay (Fig. 2C). In agreement with the idea that domain A provides a determinant for POU domain interaction, its deletion essentially abrogated the ability of OCA-B to form the ternary complex (compare lane 4, Mut. A, to lane 2, Wt). This observation suggests that the coactivation defect of mutant A is due largely to its lack of an upstream interaction with Oct-1. In contrast, the B region mutation actually enhanced the ability of OCA-B to bind to the DNA–POU-1 complex (compare lane 3, Mut. B, to lane 2, Wt). The reason for the enhancement is not known. (However, given that the combined A/B mutant protein [lane 5] showed a residual level of ternary complex formation, one likely scenario [also offered in the legend to Fig. 2C] is that the domain B mutation results in a conformational change that can increase the POU–OCA-B interaction even in the absence of domain A. Such a conformational change may also contribute, along with the loss of acidic residues per se, to the coactivation defect of the B mutant protein.) Nevertheless, given that the B mutant protein maintains the ability to supershift an octamer–POU-1 complex, its coactivation defect most likely originates from lack of a downstream interaction(s). By virtue of a lack of both interactions, it is reasonable that the A/B double mutant protein shows an even more reduced coactivation capacity (Fig. 2A and B).
To further establish these ideas, dominant negative potentials of wild-type versus mutant OCA-B proteins were tested. Namalwa cell (Nam; a B-cell lymphoma line) nuclear extract was supplemented with recombinant proteins at a 6:1 molar ratio of exogenous to endogenous OCA-B, and the IgH promoter activity was determined in an in vitro transcription assay (Fig. 2D). As predicted, the B mutant protein showed a significant dominant negative effect (compare lane 3 to lane 1), in agreement with the idea that it can bind strongly to the IgH promoter–Oct-1 complex and interfere, competitively, with the binding of endogenous OCA-B. In contrast, the A and A/B mutant proteins had no (or marginal) effects (compare lanes 4 and 5 to lane 1), whereas exogenously added wild-type OCA-B slightly boosted IgH promoter activity (compare lane 2 to lane 1). The inability of the A mutant protein, which maintains the normal domain B (Fig. 1A), to exert a dominant negative effect suggests that OCA-B may interact with a downstream target(s) more strongly when it is on the promoter than when it is free in solution.
Mechanism of action of OCA-B: essential role for PC4 and synergism with PC2 in a purified reconstituted system.
Because OCA-B alone can confer B-cell level activation on Ig promoters, its function has been analyzed mainly in crude HeLa nuclear extracts (see above and references 30 and 31). However, an earlier study (31) indicated that it can also function in a system reconstituted with Oct-1, highly purified HeLa-derived basal factors and RNA polymerase II, and general coactivator fraction USA. This suggested that Ig promoter activation by the specialized coactivator OCA-B requires still other general (USA-derived) cofactors (reviewed in reference 18). In the case of transcription activation by Gal4-based activators on model promoters with multiple Gal4 sites, the USA-derived positive cofactor PC4 can replace, and is as potent as, the complete USA fraction (e.g., 11, 22; reviewed in reference 18). However, our earlier study showed that PC4 alone could not substitute for USA in mediating Oct-1/OCA-B function in a reconstituted system (31), thus raising the question of whether the Gal4-based artificial system had “short-circuited” a regulatory pathway normally requiring more than one component of the USA fraction.
To gain further insights into this question, the first objective was to determine whether PC4 is, indeed, essential for the ability of USA to mediate activation by Oct-1 and OCA-B. To this end, PC4 was immunodepleted from the USA fraction (Fig. 3A) and the ability of the PC4-deficient USA to support OCA-B function with Oct-1 was assessed (Fig. 3B). The PC4-free USA was found to be almost completely inactive in this system (compare lane 7 to lane 6), suggesting that PC4 is, indeed, an essential USA component for Oct-1 and OCA-B function in the reconstituted system. The transcription level was almost fully restored by addition of recombinant PC4 protein (lane 8). Given that PC4 alone could not replace the USA fraction to support Oct-1 and OCA-B function (31), we conclude that it must act synergistically with another component(s) in the USA fraction.
To identify the component(s) that acts in conjunction with PC4, native and PC4-depleted USA fractions were further fractionated. An activity chromatographically identical to PC2 (24) was found to be jointly required with the recombinant PC4 to fully support the Oct-1- and OCA-B-dependent activation of the IgH promoter (Fig. 3C, lanes 7 and 8), whereas a fraction containing PC1 and PC3, in addition to PC4, was inactive (data not shown). As can be seen, appropriate titration of both PC4 and PC2 produced a transcription level equivalent to that obtained with the intact USA fraction (compare lane 7 to lane 2). On the other hand, and in agreement with prior studies of PC4 (31), the IgH promoter activity was compromised in the transcription system containing either PC4 or PC2 alone (compare lanes 3 to 6 to lanes 2, 7, and 8). Increasing the level of PC2 did not boost the transcription and, in agreement with the results of earlier studies (11, 22), increasing the level of PC4 eventually repressed the transcription system (data not shown). Taken together, these results document a synergism between PC4 and PC2 that is critical for optimal Oct-1 and OCA-B function in the reconstituted system. Note that the present analysis, in contrast to that in our previous study describing the cloning of OCA-B (31), used increased amounts of promoter template DNA, as well as larger amounts of various transcription factors, to maximize the promoter activity such that, while maintaining 8- to 10-fold coactivation by OCA-B, the level of IgH transcription in the reconstituted system is more comparable to that observed in a crude system. The considerable activation by PC4 alone observed here (Fig. 3C, compare lanes 4 and 6 to lane 1) contrasts with the previous report showing almost no activation by PC4 (Fig. 8 in reference 31) but may reflect either genuine coactivation by PC4 intrinsic to this particular redefined system or, given the observed PC2-PC4 synergism, minor PC2 contamination in the partially purified TFIID and the TFIIE/F/H fractions that were added at higher levels to accommodate a higher template dose. Because PC2 is still poorly defined, we cannot distinguish these two possibilities. Nonetheless, the PC4-PC2 synergism was clearly evident in this experiment.
Identification of PC4 as a downstream target for OCA-B function.
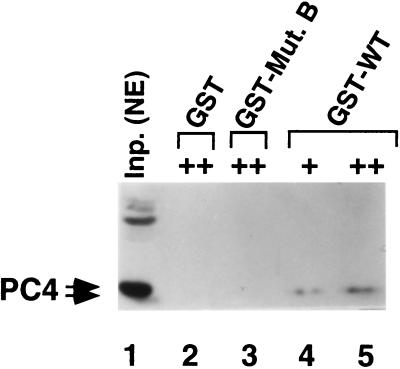
As mentioned earlier, the defect of the B mutant form of OCA-B is due to loss of a downstream interaction. This prompted us to search for an OCA-B downstream target(s). PC4 seemed a likely target in view of its essential role in the function of OCA-B in a reconstituted system (see above). To investigate this possibility, GST fusion proteins containing residues 171 to 256 of either wild-type OCA-B or the B mutant protein were employed in pull-down assays to search for a nuclear extract factor(s) capable of interacting with the OCA-B activation domain. As shown in the immunoblot analysis of Fig. 4, the functional (nonphosphorylated) form of PC4 bound to a wild-type OCA-B–GST fusion protein but not to the mutant B-GST fusion protein (or GST alone) (compare lanes 4 and 5 to lanes 2 and 3). This result is consistent with a direct role for PC4 in mediating OCA-B function through the OCA-B activation domain. Purified recombinant PC4 bound to OCA-B in the same fashion (data not shown), suggesting that the OCA-B–PC4 interaction is direct.
FIG. 4.
Identification of PC4 as a potential downstream target for the function of OCA-B. The rationale for this experiment is explained in the text, and experimental details are described in Materials and Methods. Note that only a portion (residues 171 to 256) of wild-type (WT) OCA-B and the B mutant (Mut.) protein was fused to GST. + and ++ represent two (lower and higher, respectively) concentrations of immobilized GST or GST fusions. Inp. NE, input nuclear extract.
A redundant activity for PC4 function in a crude system.
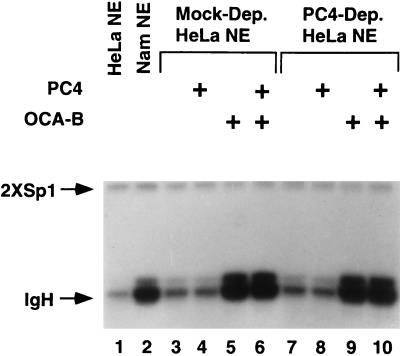
Given that OCA-B can function in crude HeLa nuclear extracts to activate the Ig promoters, it was of interest to explore a possible role of PC4 in the coactivation pathway in the crude transcription system on the grounds that it may contain a more complete complement of nuclear factors. To this end, the PC4 protein was quantitatively removed by immunodepletion from a HeLa nuclear extract (Fig. 3A, right panel) and the PC4-depleted extract was analyzed for the ability to transcribe the Oct-1/OCA-B-dependent IgH promoter and a control Sp1-dependent promoter (Fig. 5). Surprisingly, the PC4-deficient nuclear extract (lanes 7 to 10) was as potent as the mock-depleted nuclear extract (lanes 3 to 6) in transcribing both templates and OCA-B functioned equally well in PC4+ and PC4− extracts to stimulate the IgH promoter (compare lanes 5 and 6 to lanes 9 and 10) to a level similar to that observed in a B-cell extract (lane 2). These results suggest the presence of a redundant activity capable of compensating for the loss of PC4 in the crude nuclear extract. Such an activity is likely missing from the chromatographically purified USA fraction, making PC4 an essential component for the function of Oct-1/OCA-B in the reconstituted system.
FIG. 5.
PC4 depletion (Dep.) from a nuclear extract (NE) does not affect the transcription of either octamer/Oct-dependent (IgH) or -independent (2×Sp1) promoters, suggesting a potential redundant activity for PC4 in the nuclear extracts. Untreated HeLa (lane 1), Namalwa (B cell; lane 2), mock-depleted HeLa (lanes 3 to 6), and PC4-depleted HeLa nuclear extracts were used to transcribe the promoters in the absence or the presence (+) of PC4 (50 ng) and/or OCA-B (20 ng), as indicated. While the transcription level of the control template (2×Sp1) is constant in all lanes, the IgH promoter responds to OCA-B in either mock-depleted (lanes 5 and 6 versus lanes 3 and 4) or PC4-depleted (lanes 9 and 10 versus lanes 7 and 8) HeLa nuclear extracts to a level similar to that observed in the B-cell extract (compare lanes 5 and 6 and lanes 9 and 10 to lane 2). The control template (2×Sp1) is described in reference 42.
DISCUSSION
In the present study, targeted mutagenesis, in conjunction with in vitro and in vivo assays, has been used to define domains of the B-cell-specific coactivator OCA-B that are critical for its proper function with the DNA binding activator Oct-1. Further investigations aimed at gaining more insights into the mechanism of action of OCA-B revealed a synergism between general coactivators PC2 and PC4 in mediating the activation or coactivation by Oct-1/OCA-B and an OCA-B–PC4 interaction that is of apparent functional relevance. Finally, studies with less purified systems revealed a functional redundancy in general coactivators. Taken together, these studies lead to a refined model of Oct-1/OCA-B function that appears to parallel that of some viral coactivators.
OCA-B domains for recruitment to the promoter.
An N-terminal OCA-B domain that shares weak similarity with an E1A region was implicated in Ig promoter targeting through interaction with the Oct-1 POU domain. However, it appears that OCA-B contacts surfaces of both the POU-specific and POU-homeo subdomains of Oct-1 (2, 28), as well as nucleotides in the octamer element (2, 4), suggesting that several determinants in OCA-B may act in concert to form a stable DNA-activator-coactivator ternary complex. Supporting this notion is the observation that several OCA-B residues in a region about 20 aa C terminal to domain A (Fig. 1) were shown to be critical for ternary complex formation (14). Because the Oct-1 POU-specific and POU-homeo subdomains contact opposite sides of the DNA double helix (20), the octamer–POU-1–OCA-B ternary complex may form a highly stable ringlike structure important for high level, long-term Ig promoter-enhancer function in activated B cells.
An OCA-B domain involved in promoter activation and general coactivator requirements for its function.
The acidic OCA-B region (domain B in Fig. 1) was shown to be critical for coactivator function (Fig. 2A and B) but not for binding to Oct-1 or an Oct-1–DNA complex (Fig. 2C). Further confirming this notion is the fact that while a Gal4(1-94)–OCA-B fusion protein functions as an activator on promoters containing Gal4 binding sites (13, 53), a mutant with a deletion encompassing domain B loses the activation capability (53). Protein-protein interaction assays also suggest that domain B of OCA-B provides a determinant for the interaction with the general coactivator PC4 (Fig. 4). Given its ability to interact with wild-type OCA-B but not the mutant B version, PC4 appears to be a genuine downstream target for the function of OCA-B. This is in accordance with the essential role for PC4, and its synergism with general coactivator PC2, in supporting the optimal function of Oct-1/OCA-B in a reconstituted system. Since PC4 alone is as potent as the USA fraction in mediating the function of Gal4-based artificial activators on synthetic templates with multiple Gal4-binding sites in the same reconstituted system (e.g., reference 11), the PC4-PC2 synergism that was observed for the function of Oct-1/OCA-B may reflect the use of coactivators in the more physiological context of a natural activator and promoter. It is relevant to note that USA was originally identified as a crude coactivator fraction capable of mediating high levels of activation by natural activators Sp1, USF (34), and NFκB (23), and, at least for Sp1-dependent activation, USA-derived fractions were not as active as partially purified USA (34). A separate study (9) also has shown synergism between PC4 and PC2 in mediating activation by thyroid hormone receptor in conjunction with a ligand-dependent activating complex.
A model for Oct-1/OCA-B function.
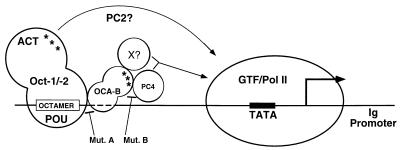
On the basis of both earlier work and the new findings shown here, we present a refined model for the function of Oct-1/OCA-B (Fig. 6). OCA-B is recruited to the target Ig promoter through primary interaction with the Oct-1 POU domain, which may precede Oct-1-DNA interactions (30, 31), and through secondary interactions with DNA (2, 4). In conjunction with Oct-1, OCA-B then provides an activation function. Given the documented physical and functional interaction between OCA-B and PC4, the OCA-B activation domain may function by influencing the basal transcription machinery with PC4 as an adapter (11). It is important to note that the defined activation domain(s) (ACT; 12, 37, 55) in the Oct factors also plays a key role in promoter activation because its removal, while not abrogating the OCA-B recruitment, is detrimental for OCA-B coactivation (31), suggesting a synergism between the activation domain in OCA-B and that in Oct-1 and -2. This synergism parallels the synergism between PC4 and PC2 that is required for the proper function of Oct-1/OCA-B in the reconstituted system, raising the intriguing possibility of a functional (or physical) interaction of PC2 and the Oct activation domain(s). Since PC2 is a potent but less well-characterized activity among the USA-derived coactivators (reviewed in reference 18), its synergism with PC4 in mediating the function of OCA-B/Oct-1 may provide a useful assay for its further characterization. A redundant activity (X in Fig. 6) is also implied by the PC4 depletion analysis in crude extracts (see below). The model also indicates the basis for the loss of function of the OCA-B A (deficiency in interaction with Oct-1 and -2) and B (deficiency in interaction with PC4, and potentially the redundant activity [X], that also leads to a dominant negative phenotype) mutant proteins.
FIG. 6.
Model for OCA-B function. Mut., mutant; GTF, general transcription factors; ACT, activation domain(s) of Oct-1 and -2; Pol II, RNA polymerase II. Asterisks denote the activation domains in either OCA-B or Oct-1 and -2. The dashed lines indicate that, in addition to contacting residues within the octamer motif (2, 4), OCA-B may also contact downstream sequences, as revealed by a footprinting assay (32). See the text for a full description of the model.
Functional analogy of OCA-B to viral coactivators.
By several criteria, OCA-B is a cellular counterpart of at least two viral coactivators, namely, the adenovirus E1A and HSV VP16 proteins. E1A is a promiscuous coactivator capable of interacting with a number of cellular DNA binding activators that include USF, Sp1, the ATF family, c-Jun (29), and Oct-4 (49) via their diverse DNA binding motifs and thus possesses the potential to regulate the transcription of multiple genes lacking a common promoter element. Of interest in relation to OCA-B is the observation that an E1A region (residues 179 to 193 in 13S E1A) implicated in promoter targeting (29) overlaps the E1A region (residues 180 to 203) that shares weak sequence similarity with OCA-B domain A (Fig. 1), which is implicated in Ig promoter targeting through interaction with the Oct-1 POU domain (Fig. 2C).
Weak (and short) sequence similarity notwithstanding, the functional analogy between OCA-B and E1A is probably best exemplified by the activation of reporter genes with an octamer-containing enhancer in embryonal carcinoma (EC) cells (49). First, in differentiated EC cells, the reporter genes can be stimulated by Oct-4 (also named Oct-3) and 13S E1A in a synergistic fashion; second, as is the case for Oct-1/OCA-B function; activation domains from both the activator (Oct-4) and the coactivator (E1A) are jointly required for optimal enhancer function; third, Oct-4, E1A, and octamer DNA form a ternary complex, as revealed by gel shift analysis; finally, in undifferentiated EC stem cells, a high level of activation can be achieved by endogenous Oct-4 alone. Given all of these parallels, it is reasonable to postulate that Oct-4 functions in undifferentiated EC stem cells with an E1A functional homolog that may be a member of a cellular coactivator family represented by OCA-B.
OCA-B also shows a remarkably close resemblance to the viral coactivator VP16 with respect to both interactions and mechanism of activation. VP16 activates HSV immediate-early genes by engaging in the formation of a multiprotein-promoter complex in conjunction with Oct-1 and another host cell factor (reviewed in references 5 and 15). Like OCA-B, VP16 interacts with promoter-bound Oct-1 through the POU domain (e.g., references 26 and 43) and has both a DNA binding activity contingent on Oct-1 binding to DNA (25, 52; reviewed in references 5 and 15) and an acidic activation domain that has the potential to interact with the general cofactor PC4 (11). The observation that, in both cases, mutations in the activation domains either severely weaken or abolish the ability to interact with PC4 (11 and this work) further supports the functional relevance of a PC4-activation domain interaction. The function and direct interaction of PC4 with several defined acidic activation domains, including those of AH, IE, E1A, VP16 (11), and OCA-B (this work) strongly argue for the role of PC4 as an adapter that bridges promoter-bound transcription factors and cofactors to the basal transcriptional machinery by interacting with TFIIA (11) and/or RNA polymerase II (33). Finally, given the recent identification of a specialized Oct-1 cofactor activity involved in the S-phase activation of an octamer-dependent H2B promoter (32), it is probable that at least some cellular genes are controlled by gene-specific coactivators that function analogously to viral coactivators and are represented by OCA-B.
Redundancy in general coactivator function.
In sharp contrast to the observation that PC4 is essential for the Oct-1/OCA-B function in a reconstituted system, PC4 is dispensable for the function of Oct-1/OCA-B in a crude (nuclear extract) system (Fig. 5). This suggests the existence of a redundant activity that is missing in the purified components used in the reconstituted assay system. This redundant activity (X in Fig. 6) may serve as an alternative adapter capable of mediating the OCA-B–general transcription factor interaction, thus bypassing the requirement for PC4 in a crude transcription system. Indeed, a potential candidate for such an activity has been detected by purification methods (60). Functional redundancy could be a hallmark of regulatory pathways in transcription. Thus, despite an absolute requirement of TATA box binding protein-associated factor (TAFS) for activator function in purified reconstituted systems (reviewed in, e.g., reference 57), studies with yeast (conditional knockout; 1, 36, 59) and less-purified human cell-free systems (40) challenge the view of a universal role of TAFs in promoter activation and imply the existence of a distinct class of functionally redundant activities (e.g., those present in the RNA polymerase II holoenzyme [reviewed in references 3, 21, and 38]) that may act through distinct mechanisms. As is the case for the function of OCA-B, this implies alternative pathways for communication between activators and/or coactivators and the basal transcription machinery.
ACKNOWLEDGMENTS
Y.L. thanks M. Kretzschmar and S. Malik for generous gifts of general factors and cofactors used in pilot experiments. We also thank other members of the Roeder laboratory for encouragement and stimulating discussions.
This work was funded by NIH grant CA43567 and by a Johnson and Johnson Focused Giving Award.
REFERENCES
- 1.Apone L M, Virbasius C A, Reese J C, Green M R. Yeast TAFII90 is required for cell-cycle progression through G2/M but not for general transcription activation. Genes Dev. 1996;10:2368–2380. doi: 10.1101/gad.10.18.2368. [DOI] [PubMed] [Google Scholar]
- 2.Babb R, Cleary M A, Herr W. OCA-B is a functional analog of VP16 but targets a separate surface of the Oct-1 POU domain. Mol Cell Biol. 1997;17:7295–7305. doi: 10.1128/mcb.17.12.7295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Björlund S, Kim Y-J. Mediator of transcriptional regulation. Trends Biochem Sci. 1996;21:335–337. doi: 10.1016/s0968-0004(96)10051-7. [DOI] [PubMed] [Google Scholar]
- 4.Cepek K L, Chasman D I, Sharp P A. Sequence-specific DNA binding of the B-cell-specific coactivator OCA-B. Genes Dev. 1996;10:2079–2088. doi: 10.1101/gad.10.16.2079. [DOI] [PubMed] [Google Scholar]
- 5.Cleary M A, Stern S, Tanaka M, Herr W. Differential positive control by Oct-1 and Oct-2: activation of a transcriptionally silent motif through Oct-1 and VP16 corecruitment. Genes Dev. 1993;7:72–83. doi: 10.1101/gad.7.1.72. [DOI] [PubMed] [Google Scholar]
- 6.Corcoran L, Karvelas M. Oct-2 is required early in T cell-independent B cell activation for G1 progression and for proliferation. Immunity. 1994;1:635–645. doi: 10.1016/1074-7613(94)90035-3. [DOI] [PubMed] [Google Scholar]
- 7.Corcoran L, Karvelas M, Nossal G, Ye Z, Jacks T, Baltimore D. Oct-2, although not required for early B-cell development, is critical for later B-cell maturation and for postnatal survival. Genes Dev. 1993;7:570–582. doi: 10.1101/gad.7.4.570. [DOI] [PubMed] [Google Scholar]
- 8.Feldhaus A, Klug C, Arvin K, Singh H. Targeted disruption of the oct-2 locus in a B cell provides genetic evidence for two distinct cell type-specific pathways of octamer element-mediated gene activation. EMBO J. 1993;12:2763–2772. doi: 10.1002/j.1460-2075.1993.tb05937.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fondell, J., M. Guermah, and R. G. Roeder. Unpublished observations.
- 10.Ge H, Martinez E, Chiang C M, Roeder R G. Activator-dependent transcription by mammalian RNA polymerase II: in vitro reconstitution with general transcription factors and cofactors. Methods Enzymol. 1996;272:57–71. doi: 10.1016/s0076-6879(96)74008-9. [DOI] [PubMed] [Google Scholar]
- 11.Ge H, Roeder R G. Purification, cloning, and characterization of a human coactivator, PC4, that mediates transcriptional activation of class II genes. Cell. 1994;78:513–523. doi: 10.1016/0092-8674(94)90428-6. [DOI] [PubMed] [Google Scholar]
- 12.Gerster T, Balmaceda C-G, Roeder R G. The cell type-specific octamer transcription factor OTF-2 has two domains required for the activation of transcription. EMBO J. 1990;9:1635–1643. doi: 10.1002/j.1460-2075.1990.tb08283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gstaiger M, Knoepfel L, Georgiev O, Schaffner W, Hovens C M. A B-cell coactivator of octamer-binding transcription factors. Nature. 1995;373:360–362. doi: 10.1038/373360a0. [DOI] [PubMed] [Google Scholar]
- 14.Gstaiger M, Georgiev O, van Leeuwen H, van der Vliet P, Schaffner W. The B cell coactivator Bob1 shows DNA sequence-dependent complex formation with Oct-1/Oct-2 factors, leading to differential promoter activation. EMBO J. 1996;15:2781–2790. [PMC free article] [PubMed] [Google Scholar]
- 15.Herr W, Cleary M A. The POU domain: versatility in transcriptional regulation by a flexible two-in-one DNA-binding domain. Genes Dev. 1995;9:1679–1693. doi: 10.1101/gad.9.14.1679. [DOI] [PubMed] [Google Scholar]
- 16.Herr W, Sturm R A, Clerc R G, Corcoran L M, Baltimore D, Sharp P A, Ingraham H A, Rosenfeld M G, Finney M, Ruvkun G, Horvitz H R. The POU domain: a large conserved region in the mammalian pit-1, oct-1, oct-2, and Caenorhabditis elegans unc-86 gene products. Genes Dev. 1988;2:1513–1516. doi: 10.1101/gad.2.12a.1513. [DOI] [PubMed] [Google Scholar]
- 17.Hoffmann A, Roeder R G. Purification of his-tagged proteins in non-denaturing conditions suggests a convenient method for protein interaction studies. Nucleic Acids Res. 1991;19:6337–6338. doi: 10.1093/nar/19.22.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaiser K, Meisterernst M. The human general co-factors. Trends Biochem Sci. 1996;21:343–345. [PubMed] [Google Scholar]
- 19.Kim U, Qin X-F, Gong S, Stevens S, Luo Y, Nussenzweig M C, Roeder R G. The B-cell-specific transcription coactivator OCA-B/OBF-1/Bob-1 is essential for the normal production of immunoglobulin isotypes. Nature. 1996;383:543–547. doi: 10.1038/383542a0. [DOI] [PubMed] [Google Scholar]
- 20.Klemm J D, Rould M A, Aurora R, Herr W, Pabo C O. Crystal structure of the Oct-1 POU domain bound to an octamer site: DNA recognition with tethered DNA binding modules. Cell. 1994;77:21–32. doi: 10.1016/0092-8674(94)90231-3. [DOI] [PubMed] [Google Scholar]
- 21.Koleske A J, Young R A. The RNA polymerase II holoenzyme and its implications for gene regulation. Trends Biochem Sci. 1995;20:113–116. doi: 10.1016/s0968-0004(00)88977-x. [DOI] [PubMed] [Google Scholar]
- 22.Kretzschmar M, Kaiser K, Lottspeich F, Meisterernst M. A novel mediator of class II gene transcription with homology to viral immediate-early transcriptional regulators. Cell. 1994;78:525–534. doi: 10.1016/0092-8674(94)90429-4. [DOI] [PubMed] [Google Scholar]
- 23.Kretzschmar M, Meisterernst M, Scheidereit C, Li G, Roeder R G. Transcriptional regulation of the HIV-1 promoter by NF-κB in vitro. Genes Dev. 1992;6:761–774. doi: 10.1101/gad.6.5.761. [DOI] [PubMed] [Google Scholar]
- 24.Kretzschmar M, Stelzer G, Roeder R G, Meisterernst M. RNA polymerase II cofactor PC2 facilitates activation of transcription by Gal4-AH in vitro. Mol Cell Biol. 1994;14:3927–3937. doi: 10.1128/mcb.14.6.3927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kristie T M, Sharp P A. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV α trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 26.Lai J-S, Cleary M A, Herr W. A single amino acid exchange transfers VP16-induced positive control from the Oct-1 to the Oct-2 homeodomain. Genes Dev. 1992;6:2058–2065. doi: 10.1101/gad.6.11.2058. [DOI] [PubMed] [Google Scholar]
- 27.Landolfi N F, Capra J D, Tucker P W. Interaction of cell-type-specific nuclear proteins with immunoglobulin VH promoter region sequences. Nature. 1986;323:548–551. doi: 10.1038/323548a0. [DOI] [PubMed] [Google Scholar]
- 28.Li, H., Y. Luo, and R. G. Roeder. Unpublished observations.
- 29.Liu F, Green M R. Promoter targeting by adenovirus E1a through interaction with different cellular DNA-binding domains. Nature. 1994;368:520–525. doi: 10.1038/368520a0. [DOI] [PubMed] [Google Scholar]
- 30.Luo Y, Fujii H, Gerster T, Roeder R G. A novel B cell-derived coactivator potentiates the activation of immunoglobulin promoters by octamer-binding transcription factors. Cell. 1992;71:231–241. doi: 10.1016/0092-8674(92)90352-d. [DOI] [PubMed] [Google Scholar]
- 31.Luo Y, Roeder R G. Cloning, functional characterization, and mechanism of action of the B-cell-specific transcriptional coactivator OCA-B. Mol Cell Biol. 1995;15:4115–4124. doi: 10.1128/mcb.15.8.4115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo, Y., and R. G. Roeder. Unpublished observations.
- 33.Malik S, Guermah M, Roeder R G. A dynamic model for PC4 coactivator function in RNA polymerase II transcription. Proc Natl Acad Sci USA. 1998;95:2192–2197. doi: 10.1073/pnas.95.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meisterernst M, Roy A, Lieu H-M, Roeder R G. Activation of class II gene transcription by regulatory factors is potentiated by a novel activity. Cell. 1991;66:981–993. doi: 10.1016/0092-8674(91)90443-3. [DOI] [PubMed] [Google Scholar]
- 35.Mitchell P J, Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA-binding proteins. Science. 1989;245:371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- 36.Moqtaderi Z, Bai Y, Poon D, Weil P A, Struhl K. TBP-associated factors are not generally required for transcription activation in yeast. Nature. 1996;383:188–190. doi: 10.1038/383188a0. [DOI] [PubMed] [Google Scholar]
- 37.Müller-Immerglück M M, Schaffner W, Matthias P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 1990;9:1625–1634. doi: 10.1002/j.1460-2075.1990.tb08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Myers L C, Gustafsson C M, Bushnell D A, Lui M, Erdjument-Bromage H, Tempst P, Kornberg R D. The Med proteins of yeast and their function through the RNA polymerase II carboxy-terminal domain. Genes Dev. 1997;12:45–54. doi: 10.1101/gad.12.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nielsen P J, Georgiev O, Lorenz B, Schaffner W. B lymphocytes are impaired in mice lacking the transcriptional co-activator Bob1/OCA-B/OBF1. Eur J Immunol. 1996;26:3214–3218. doi: 10.1002/eji.1830261255. [DOI] [PubMed] [Google Scholar]
- 40.Oelgeschläger T, Tao Y, Kang Y-K, Roeder R G. Transcription activation via enhanced preinitiation complex assembly in a human cell free system lacking TAFIIs. 1998. Mol. Cell, in press. [DOI] [PubMed] [Google Scholar]
- 41.Pierani A, Heguy A, Fujii H, Roeder R G. Activation of octamer-containing promoters by either octamer binding transcription factor 1 (OTF-1) or OTF-2 and requirement of an additional B-cell-specific component for optimal transcription of immunoglobulin promoters. Mol Cell Biol. 1990;10:6204–6215. doi: 10.1128/mcb.10.12.6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pognonec P, Roeder R G. Recombinant 43-kDa USF binds to DNA and activates transcription in a manner indistinguishable from that of natural 43/44-kDa USF. Mol Cell Biol. 1991;11:5125–5136. doi: 10.1128/mcb.11.10.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pomerantz J L, Kristie T M, Sharp P A. Recognition of the surface of a homeo domain protein. Genes Dev. 1992;6:2047–2057. doi: 10.1101/gad.6.11.2047. [DOI] [PubMed] [Google Scholar]
- 44.Roeder R G. The role of general initiation factors in transcription by RNA polymerase II. Trends Biochem Sci. 1996;21:327–335. [PubMed] [Google Scholar]
- 45.Rosenfeld M G. POU-domain transcription factors: pou-er-ful developmental regulators. Genes Dev. 1991;5:897–907. doi: 10.1101/gad.5.6.897. [DOI] [PubMed] [Google Scholar]
- 46.Ruvkun G, Finney M. Regulation of transcription and cell identity by POU domain proteins. Cell. 1991;64:475–478. doi: 10.1016/0092-8674(91)90227-p. [DOI] [PubMed] [Google Scholar]
- 47.Ryan A K, Rosenfeld M G. POU domain family values: flexibility, partnership, and developmental codes. Genes Dev. 1997;11:1207–1225. doi: 10.1101/gad.11.10.1207. [DOI] [PubMed] [Google Scholar]
- 48.Schöler H R. Octamania: the POU factors in murine development. Trends Genet. 1991;7:323–328. doi: 10.1016/0168-9525(91)90422-m. [DOI] [PubMed] [Google Scholar]
- 49.Schöler H R, Ciesiolka T, Gruss P. A nexus between Oct-4 and E1A: implications for gene regulation in embryonic stem cells. Cell. 1991;66:291–304. doi: 10.1016/0092-8674(91)90619-a. [DOI] [PubMed] [Google Scholar]
- 50.Schubart D B, Rolink A, Kosco-Vilbois M H, Botteri F, Mattias P. B-cell-specific coactivator OBF-1/OCA-B/Bob-1 required for immune response and germinal center formation. Nature. 1996;383:538–543. doi: 10.1038/383538a0. [DOI] [PubMed] [Google Scholar]
- 51.Staudt L M, Lenardo M J. Immunoglobulin gene transcription. Annu Rev Immunol. 1991;9:373–398. doi: 10.1146/annurev.iy.09.040191.002105. [DOI] [PubMed] [Google Scholar]
- 52.Stern S, Herr W. The herpes simplex virus trans-activator VP16 recognizes the Oct-1 homeodomain: evidence for a homeodomain recognition subdomain. Genes Dev. 1991;5:2555–2566. doi: 10.1101/gad.5.12b.2555. [DOI] [PubMed] [Google Scholar]
- 53.Stevens, S., Y. Luo, and R. G. Roeder. Unpublished observations.
- 54.Strubin M, Newell J W, Mattias P. OBF-1, a novel B cell-specific coactivator that stimulates immunoglobulin promoter activity through association with octamer-binding proteins. Cell. 1995;80:497–506. doi: 10.1016/0092-8674(95)90500-6. [DOI] [PubMed] [Google Scholar]
- 55.Tanaka M, Herr W. Differential transcriptional activation by Oct-1 and Oct-2: interdependent activation domains induce Oct-2 phosphorylation. Cell. 1990;60:375–386. doi: 10.1016/0092-8674(90)90589-7. [DOI] [PubMed] [Google Scholar]
- 56.Triezenberg S J, Kingsbury R C, Mcknight S L. Functional dissection of VP16, the transactivator of herpes simplex virus immediate early gene expression. Genes Dev. 1988;2:718–729. doi: 10.1101/gad.2.6.718. [DOI] [PubMed] [Google Scholar]
- 57.Verrijzer C P, Tjian R. TAFs mediate transcriptional activation and promoter selectivity. Trends Biochem Sci. 1996;21:338–343. [PubMed] [Google Scholar]
- 58.Walker S, Hayes S, O’Hare P. Site-specific conformational alteration of the Oct-1 POU domain-DNA complex as the basis for differential recognition by Vmw65 (VP16) Cell. 1994;79:841–852. doi: 10.1016/0092-8674(94)90073-6. [DOI] [PubMed] [Google Scholar]
- 59.Walker S S, Reese J C, Apone L M, Green M R. Transcription activation in cells lacking TAFIIs. Nature. 1996;383:185–188. doi: 10.1038/383185a0. [DOI] [PubMed] [Google Scholar]
- 60.Xiao, H., Y. Luo, and R. G. Roeder. Unpublished observations.