Figure 1.
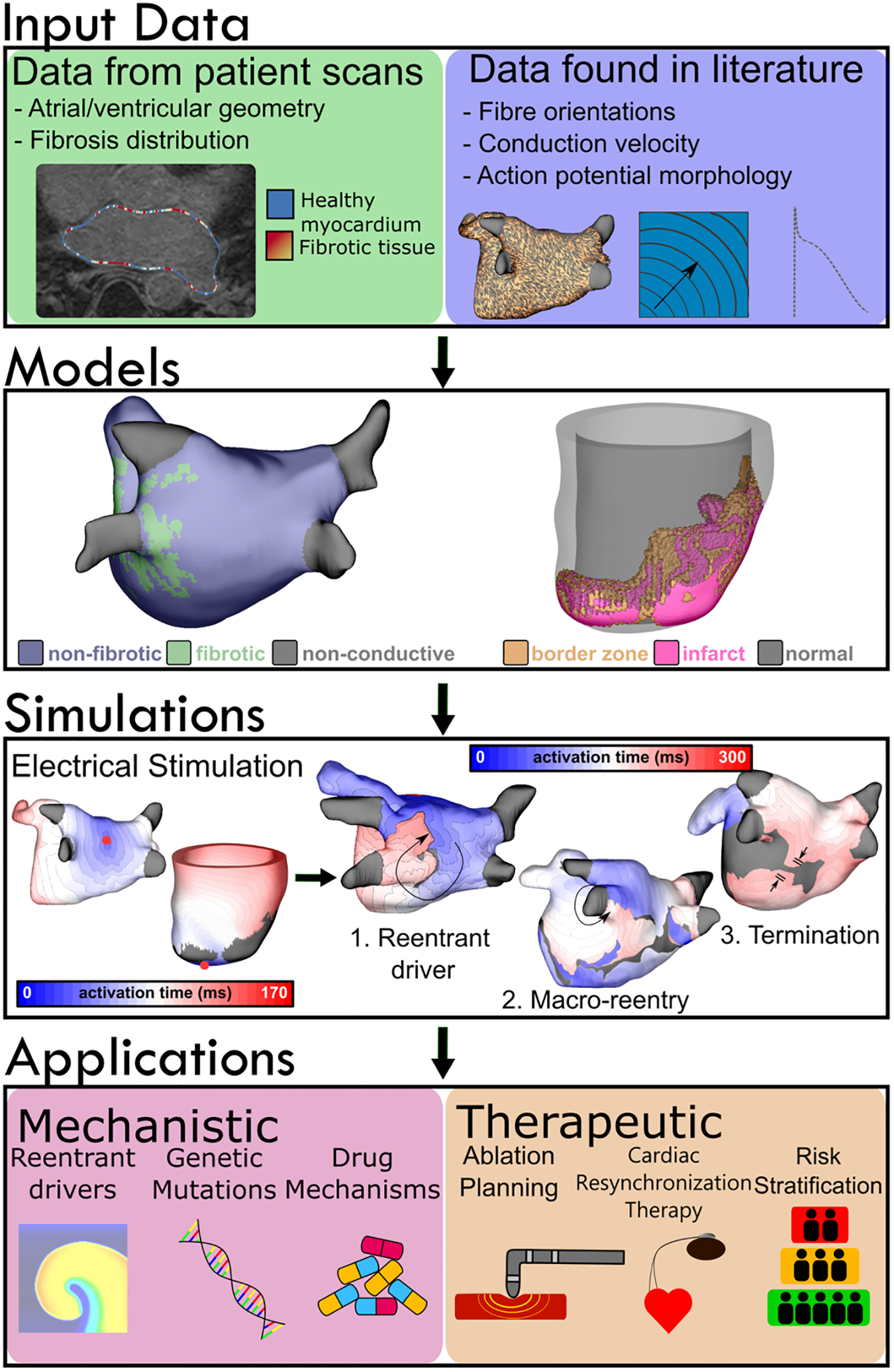
Modeling workflow for atrial and ventricular simulations. Data defining cardiac geometry and fibrosis distribution are obtained from clinical imaging modalities such as LGE-MR or computed tomography. Tissue- and cell-scale information, including action potential morphology and conduction velocity in different regions (fibrotic, non-fibrotic, gray zone, etc.) are derived from scientific literature. Fiber orientations are obtained from human atlas geometries and diffeomorphically mapped into patient-specific geometries. These components are used to assemble organ-scale models with patient-derived representations of the regional distribution of disease-related remodeling, in which electrophysiological properties are perturbed in a generic manner. Virtual electrical stimuli can be applied to any area within the model to elicit (1) reentrant drivers, (2) macro-reentry around non-conductive boundaries (e.g., around the LIPV, as shown here) or (3) termination of arrhythmic activity, indicating a return to sinus rhythm. Simulations have a range of applications from mechanistic (e.g., elucidating factors underlying reentry or anti-arrhythmic drug action) to therapeutic (e.g., ablation planning procedures or arrhythmia risk stratification).
