Extended Data Fig. 1 |. CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP Cryo-EM analysis.
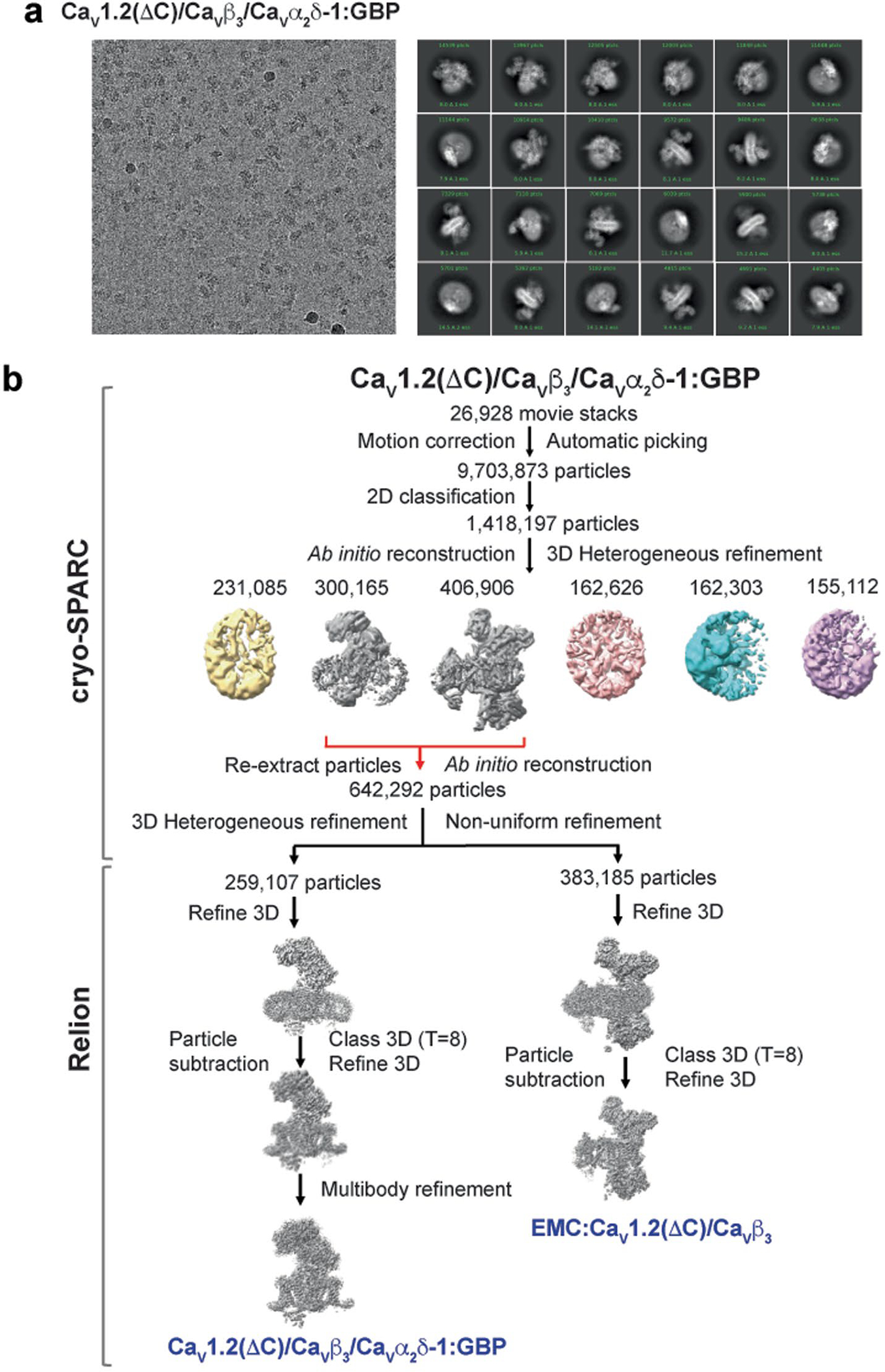
a, Exemplar CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP electron micrograph (~105,000x magnification) and 2D class averages. N = 3. b, Workflow for electron microscopy data processing for CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP sample. Initial cryoSPARC-3.2 Ab initio reconstruction identified a population of particles containing the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1 and EMC:CaV1.2(ΔC)/CaVβ3 complexes, similar to prior studies13. Red arrows indicate the two classes that were re-extracted, subjected to multiple rounds of 3D heterogeneous classification, and exported from cryoSPARC-3.2 for further 3D refinement in RELION-3.1. Particle subtraction was performed for both the refined maps in Relion-3.1 followed by 3D classification with single class and 3D refinement to get the final consensus maps. Multibody refinement was performed to enhance features of CaVα2δ-1, which was used for the CaV1.2(ΔC)/CaVβ3/CaVα2δ-1:GBP composite map. The composite map was used for model building and refinement.
