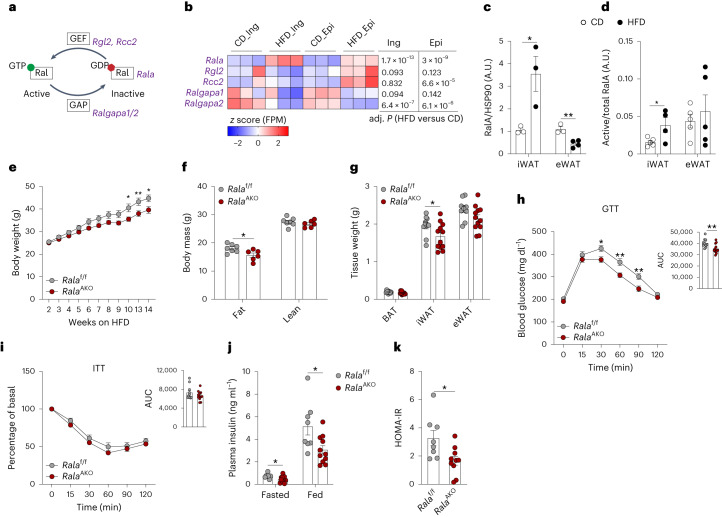
Fig. 1. White adipocyte-specific Rala deletion protects mice from high-fat-diet-induced obesity.
a, Scheme illustrating RalA activation network involving genes encoding RalA, GEF and GAP. b, RNA-seq analysis of primary inguinal (Ing) and epididymal (Epi) mature adipocytes isolated from mice (n = 3) under 16-week HFD feeding. Heat map displays transcriptional expression as z-scored FPM values. Adjusted P (adj. P) values are indicated and considered significant with values <0.05. c, Quantification of RalA protein content in mature adipocytes from iWAT and eWAT of mice fed with CD (n = 3) or HFD (n = 4) for 16 weeks. iWAT P = 0.033 CD versus HFD, eWAT P = 0.005 CD versus HFD. A.U., arbitrary units. d, Quantification of RalA GTPase activity in iWAT and eWAT of mice (n = 4) fed with CD or HFD for 4 weeks. iWAT P = 0.0448 CD versus HFD. e, Body weight of Ralaf/f (n = 8) and RalaAKO (n = 10) mice fed with 60% HFD. Longitudinal graph, P = 0.0158, P = 0.009, P = 0.0106. f, Body mass of Ralaf/f (n = 7) and RalaAKO (n = 6) mice fed with HFD for 12 weeks. Fat mass P = 0.0252. g, Fat depot weights of Ralaf/f (n = 10) and RalaAKO (n = 12) mice fed with HFD for 12 weeks. iWAT P = 0.0465. h, GTT on 11-week HFD-fed Ralaf/f (n = 10) and RalaAKO (n = 13) mice, P = 0.0174, P = 0.0036, P = 0.0069; the area under the curve (AUC) was calculated from longitudinal charts, P = 0.0062. i, ITT on 12-week HFD-fed Ralaf/f (n = 10) and RalaAKO (n = 12) mice; AUC was calculated from longitudinal chart. j, Plasma insulin levels in 8-week HFD-fed Ralaf/f and RalaAKO mice (n = 11). Fasted P = 0.0166. Fed P = 0.0329. k, HOMA-IR was calculated using fasting glucose and insulin levels from 8-week HFD-fed Ralaf/f (n = 8) and RalaAKO (n = 10) mice. P = 0.0152. Data (c–k) show mean ± s.e.m., *P < 0.05, **P < 0.01, by two-tailed Student’s t-test (c,d,f,g,j,k) or two-way analysis of variance (ANOVA) with Bonferroni’s post-test (e,h,i).