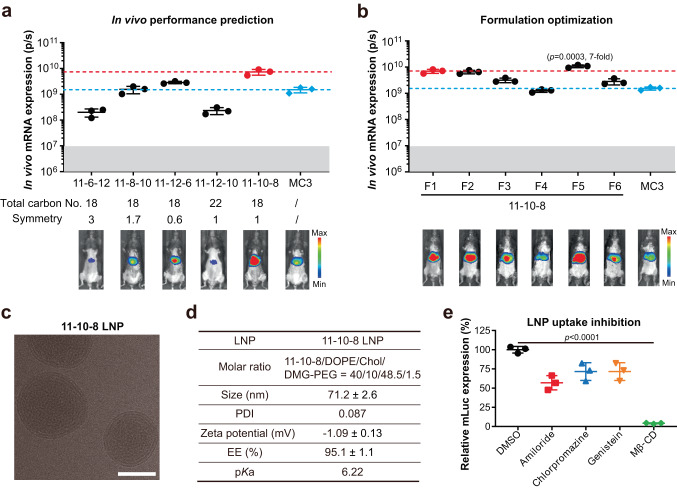
Fig. 4. Prediction, optimization, and characterization of DB-LNPs.
a Prediction and verification of in vivo performance for unidentified DB-lipidoids (n = 3 biologically independent samples). Mice were i.v. injected with mLuc-loaded LNPs at an mRNA dose of 0.1 mg/kg. BLI was performed at 4 h post-treatment and total flux was quantified. The red dashed line indicates the performance of 11-10-8 LNP, while the blue dashed line indicates the performance of MC3 LNP. The grey shadow indicates background level. b Optimization of 11-10-8 LNP formulation (n = 3 biologically independent samples). Mice were injected i.v. with mLuc-loaded LNPs at an mRNA dose of 0.1 mg/kg. BLI was performed at 4 h post-treatment and total flux was quantified. The grey shadow indicates background level. Statistical significance was evaluated by a one-way ANOVA with Tukey’s correction. c A representative cryo-EM image of 11-10-8 LNP from three independent experiments. Scale bar = 50 nm. d Physicochemical properties of 11-10-8 LNP (n = 3). e Inhibition of 11-10-8 LNP uptake by various endocytic inhibitors (n = 3 biologically independent samples). Amiloride is an inhibitor of macropinocytosis; Chlorpromazine is an inhibitor of clathrin-mediated endocytosis; Genistein is an inhibitor of caveolae-mediated endocytosis; Methyl-β-cyclodextrin (Mβ-CD) is an inhibitor of lipid raft-mediated endocytosis. Statistical significance was evaluated by a one-way ANOVA with Tukey’s correction. Data are presented as mean ± SD. Source data are provided as a Source Data file.