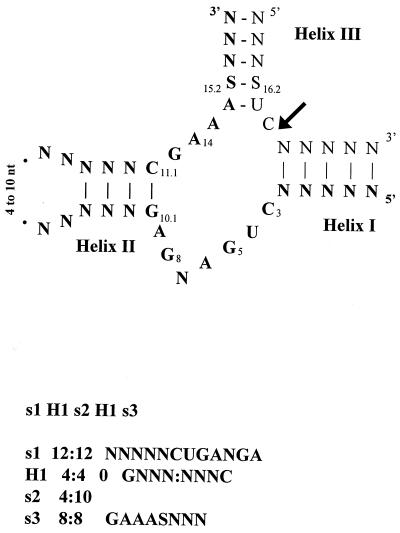
FIG. 1.
Searching the GenBank database for hammerhead ribozyme RNA domains. (Top) The secondary structure of the hammerhead domain with the conserved nucleotides indicated. The part for which the descriptor was written is shown in boldface. (Bottom) The descriptor used in the program RNAMOT (23) was composed of the following features: s1 H1 s2 H1 s3. The s1 feature scans for a 12-nucleotide (nt) sequence of the form NNNNNCUGANGA, the first 5 nucleotides of which are represented by N (any nucleotide) and which includes the required sequence CUGANGA, which is part of the conserved catalytic core. The feature H1, s2, H1 corresponds to the helix II region closed by a loop of 4 to 10 nucleotides (s2). The numbers 4:4 refer to a helix of 4 bp having zero mismatches but allowing the wobble GU. In this helix, a GC pair is required at the base. The s3 feature requires an 8-nucleotide sequence of the form GAAASNNN, where S represents C or G. This feature contains the remaining part of the conserved catalytic core and the variable nucleotides of the 3′ recognition helix. In order to catalyze a cleavage reaction, RNA defined by this descriptor must recognize a substrate RNA by base pairing. The sequence of a possible substrate is represented, and the arrow indicates the cleavage site. The numbering system is that of Hertel et al. (19).