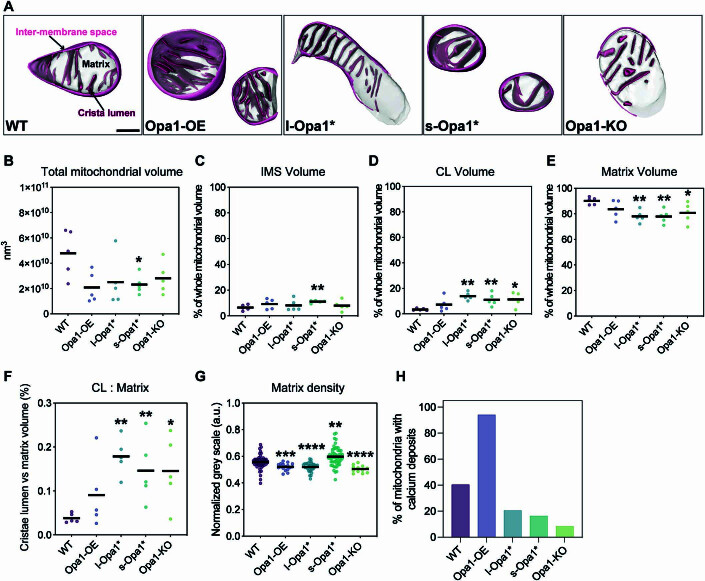
Figure EV1. Mitochondrial subcompartment volumes.
(A) Three-dimensional renderings of segmented inter-membrane space (IMS, pink surface), cristae lumen (CL, magenta surface), and matrix (translucent gray surface) volumes. Scale bar = 200 nm. (B) Total mitochondrial volume across indicated cell lines. N = 5 cells for all cell lines. (C) Quantification of IMS volume relative to total volume of each mitochondrion indicated in (B). N = 5 cells for all cell lines. (D) Quantification of CL volume relative to total volume of each mitochondrion indicated in (B). N = 5 cells for all cell lines. (E) Quantification of matrix volume relative to total volume of each mitochondrion indicated in (B). N = 5 cells for all cell lines. (F) CL to matrix ratio across cell lines. N = 5 cells for all cell lines. (G) Normalized gray scale mitochondrial matrix value across cell lines. N = 5 cells for all cell lines. (H) Graph bar representing percentage of cells with detected calcium deposits in cryo-electron tomograms. N refers to the number of mitochondria: WT = 57, Opa1-OE = 17, l-Opa1* = 39, s-Opa1* = 55, Opa1-KO = 12. Data information: Scatter plots show data distribution, the mean is shown by a bold black line. Significance of difference is tested relative to wild type using Mann–Whitney test in (B, D, E, G); *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001; and unpaired t test in (C): **p < 0.01.