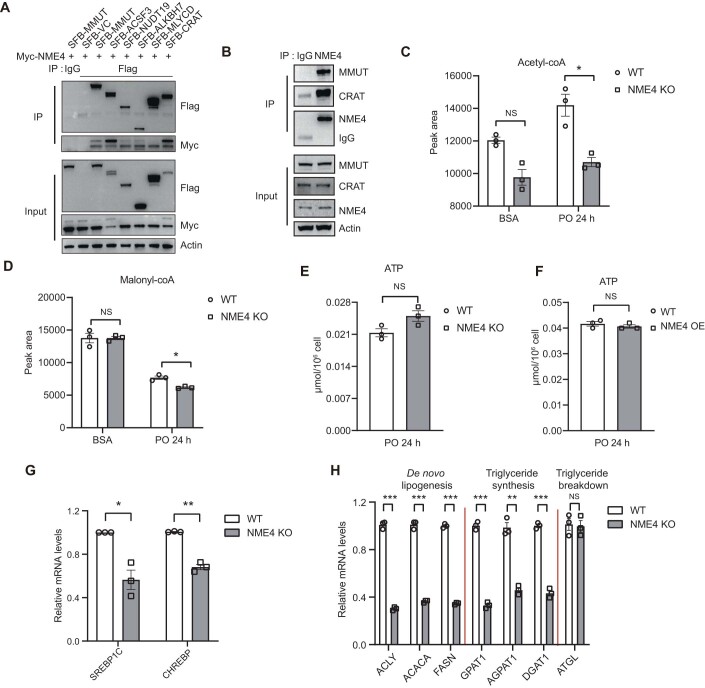
Figure EV5. NME4 regulates coenzyme A metabolism by interacting with the key enzymes in the pathway.
(A) HEK293T cells were cotransfected with Myc-tagged NME4 and C-terminal SFB (SFB)-tagged candidate genes, as indicated. The cell lysates were incubated with immunoglobulin G (IgG) control and antibodies against Flag. (B) HEK293T cell lysates were incubated with immunoglobulin G (IgG) control and antibodies against NME4. (C,D) Wild-type and NME4 KO Bel-7402 cells were treated with PO for 24 h. The acetyl-CoA (C) and malonyl-CoA (D) levels were determined by targeted metabolite analysis. Biological replicates, n = 3. (E) Wild-type and NME4 KO Bel-7402 cells were treated with PO for 24 h. ATP levels was measured by kit. Biological replicates, n = 3. (F) Wild-type and NME4-overexpression SK-Hep1 cells were treated with PO for 24 h. ATP levels was measured by kit. Biological replicates, n = 3. (G) Relative mRNA levels of SREBP1C and CHREBP were measured by RT‒qPCR. Biological replicates, n = 3. (H) Relative mRNA levels of key genes involved in de novo lipogenesis, triglyceride synthesis and triglyceride breakdown were measured by RT‒QPCR in wild-type and NME4 KO Bel-7402 cells. Biological replicates, n = 3. Data information: (C–H) data are presented as mean ± SEM. *P values ≤ 0.05, **P values ≤ 0.01, ***P values ≤ 0.001, NS -P values > 0.05 (Student’s t test, unpaired).