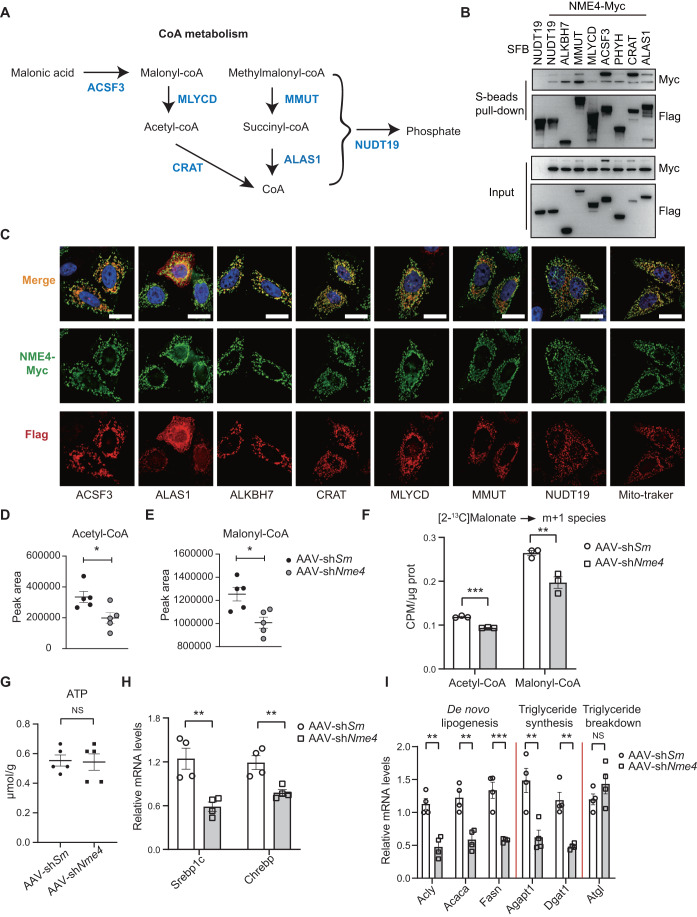
Figure 6. NME4 regulates coenzyme A metabolism by interacting with the key enzymes in the pathway.
(A) Metabolic flow chart shows the CoA-metabolic pathway and the key enzymes catalyzing CoA metabolism (KECCAM). (B,C) HEK293T cells were cotransfected with SFB-tagged KECCAM and Myc-tagged NME4. (B) The cell lysates were incubated with S beads. Five percent lysate was used as the input control. Blots probed with antibodies against the Flag and Myc epitope tags are shown. (C) The cells were subjected to immunofluorescence with an anti-Myc antibody to identify NME4 (green), an anti-Flag antibody to identify SFB-tagged KECCAM or MitoTracker (red), and DAPI (blue) and visualized by microscopy. Scale bars, 20 μm. (D,E) AAV-shScramble and AAV-shNme4 mice fed with 12 weeks HFD and livers were separated. The acetyl-CoA (D) and malonyl-CoA (E) levels were determined by targeted metabolite analysis. Biological replicates, n = 5. (F) Primary hepatocytes were treated with [2-13C] Malonate were extracted. The acetyl-CoA (m + 1) and malonyl-CoA (m + 1) levels were determined by targeted metabolite analysis. Biological replicates, n = 3. (G) AAV-shScramble and AAV-shNme4 mice fed with 12 weeks HFD and livers were separated. The ATP levels was measured by kit. Biological replicates, n = 5. (H) Relative mRNA levels of Srebp1c and Chrebp were measured by RT‒qPCR. Biological replicates, n = 4. (I) Relative mRNA levels of key genes involved in de novo lipogenesis, triglyceride synthesis, and triglyceride breakdown were measured by RT‒qPCR. Biological replicates, n = 4. Data information: (D–I) data are presented as mean ± SEM. *P values ≤ 0.05, **P values ≤ 0.01, ***P values ≤ 0.001, NS P values > 0.05 (Student’s t test, unpaired). Sm Scramble. Source data are available online for this figure.