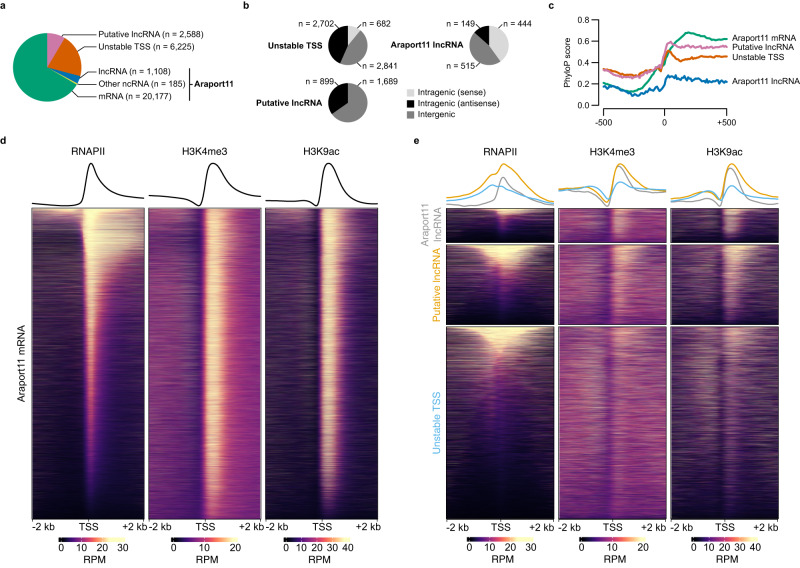
Fig. 4. Characteristics of non-coding transcription during the seed-to-seedling transition.
a Pie chart of annotated TSS types from the csRNA-seq. TSSs associated with an existing Araport11 TSS were annotated as either mRNA, lncRNA or Other ncRNA. The remaining TSSs were annotated as either Putative lncRNA (when a putative transcript could be reconstructed from the RNA-seq) or Unstable TSS (when transcript reconstruction was not possible). b Pie charts of the positional contexts of non-coding TSSs (lncRNA, Putative lncRNA and Unstable TSS). Features which did not overlap any protein coding gene were annotated as Intergenic. Features which overlapped a protein coding gene were annotated as Intragenic, and as either sense or antisense in brackets to denote the relative orientation to the overlapping gene. c Average conservation of promoters by annotated TSS type (mRNA, lncRNA, Putative lncRNA, and Unstable TSS), using PhyloP scores calculated from 63 plant species42. The coverage of scores is from 500 bp upstream and downstream of the primary TSS coordinate. d Heatmaps, and accompanying average profiles of external RNAPII110, H3K4me3115, and H3K9ac116 ChIP-seq datasets from Arabidopsis seedlings of the 4 kb area around protein coding TSSs. e A repeat of the plotting from (g), instead showing the non-coding annotated TSS types.