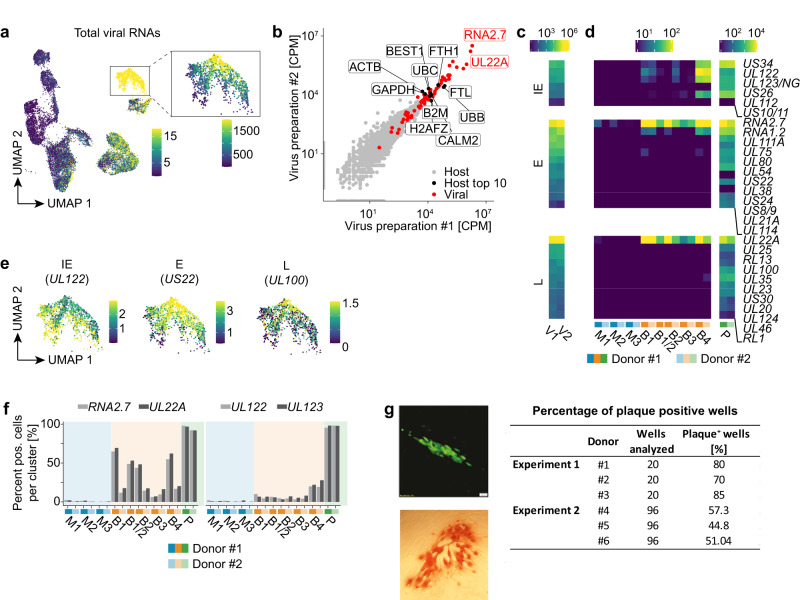
Fig. 2. Most HCMV-NG exposed moDCs contain virion-associated RNAs, but only few ones show de novo viral gene expression.
a UMAP embedding showing the total expression of viral RNAs (SCTransform normalized values) in the dataset analyzed also in Fig. 1. The color scale is chosen to highlight cells with weak viral gene expression, and all cells shown in yellow have expression values > 15. The inset shows the total expression of viral RNAs for cluster P with a different color scale resolving cells with strong viral gene expression. Here all cells shown in yellow have expression values > 1500. b Scatter plot showing the abundance of host (grey dots) and viral (red dots) RNAs (in counts per million) in the two independent virus preparations. The two most abundant viral RNAs (RNA2.7, UL22A) and the ten most abundant host RNAs (black dots) are labeled. Heatmap showing the abundance of viral RNAs in the virus preparations V1 and V2 (c) and in the clusters detected by scRNA-seq (d) (counts per million). IE genes are significantly more expressed in B4 than in B1-3 (p < 3.5e-57, two-sided Wilcoxon rank sum test). e UMAP embeddings depicting the expression (SCTransform normalized values) of representative viral RNAs of the different kinetic classes of HCMV gene expression in P. f Bar diagram showing the abundance (% of all cells that have detectable expression) of virion-associated (RNA2.7 and UL22A) and de novo transcribed (UL122 and UL123) viral RNAs in mock-treated and HCMV-NG exposed cells. g HCMV-NG exposed moDCs were FACS sorted and single NG+ moDCs were seeded on monolayers of MRC-5 cells. After 4 days of incubation, NG+ plaques were detected in the MRC-5 monolayer by wide-field fluorescent microscopy. A representative NG+ plaque is shown in the upper panel, and in the lower panel, one representative IE1+ plaque is shown, as detected by light microscopy of HRP-IE1 immunolabeled cells. The table shows the quantification of the number of wells analyzed and the number of wells showing NG+ plaques from 6 independent donors from 2 independent experiments.